Michael Perry
Research
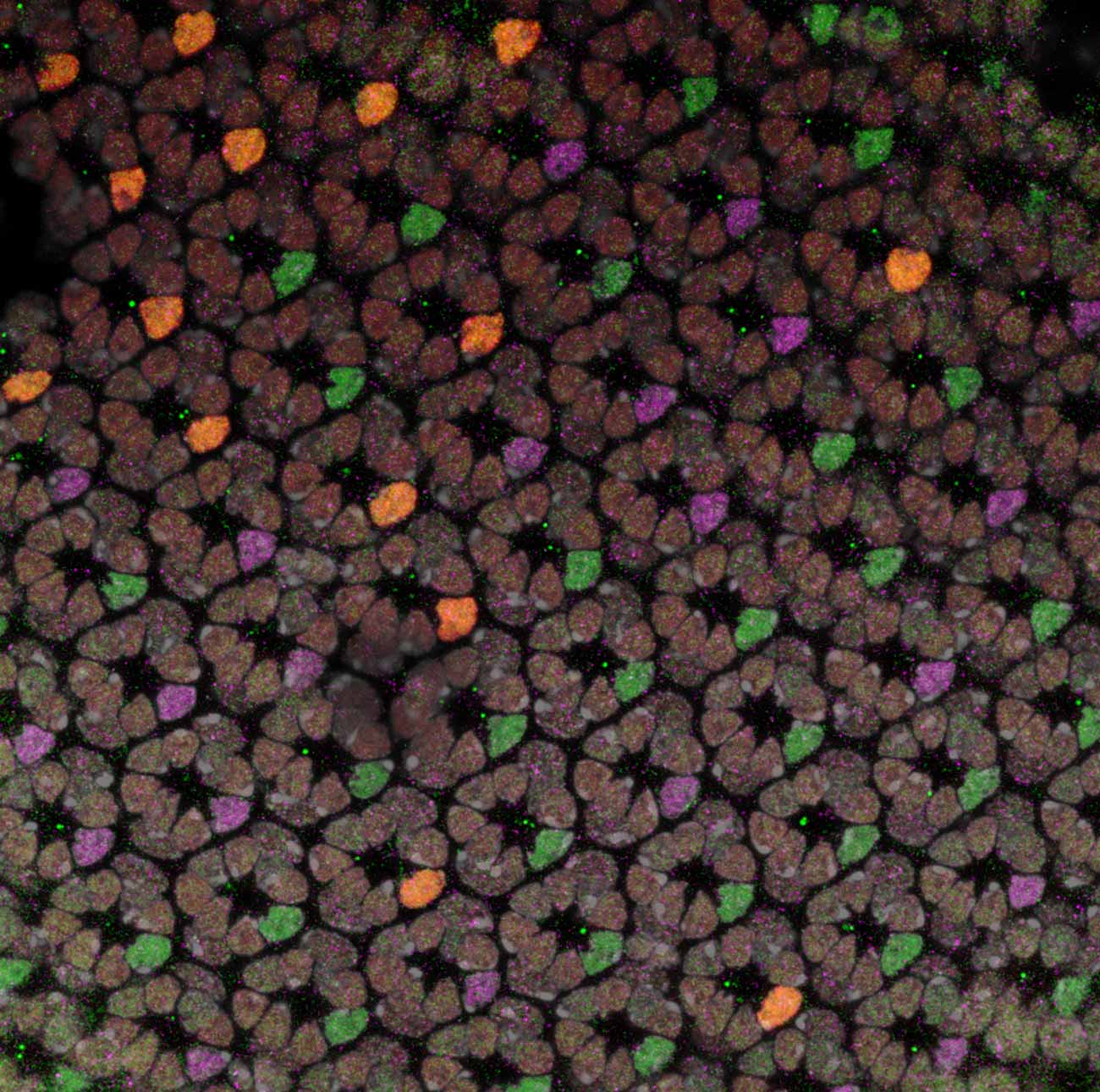
Stochastic gene expression in developing photoreceptors
One of the most interesting questions in biology is how the complexity of the nervous system is encoded in the genome: how it provides rules to build a system that can compute, remember, and produce behavioral responses. To understand which genetic changes shape neural function we leverage naturally occurring variation in nervous systems within and across species. The lab uses the insect visual system as a model, from Drosophila and other flies all the way to butterflies and wasps. Insect eyes and brains have been shaped by a rich diversity of functional requirements to exquisitely tune the nervous system and behavioral responses to allow them to thrive in an incredible range of environments.

A flower fly showing stochastic patterns on the retina
Image by Yousef Al Habshi
Natural diversity of the visual system comes in large part from changes in how and when genes are expressed during development. The advent of powerful genomic tools such as CRISPR/Cas9 and single cell sequencing have radically changed the questions we can ask in non-model systems: we can now perform targeted comparisons and functional molecular experiments in almost any species. There has never been a better time to combine approaches in model systems with experiments directly in species that exhibit interesting differences in form, function, and behavior as a targeted means of unraveling neural complexity. How are the full diversity of neural types specified, and how do new neural types evolve and “plug in” to existing circuity? How do such changes modify downstream neural function? Does the interconnected nature of the nervous system influence how it evolves? The long-term goal of the lab is to understand the genetic basis of nervous system development and evolution at levels ranging from molecules to behavior.

CRISPR in butterflies: mosaic yellow mutant Papilio xuthus and Vanessa cardui
Select Publications
- Yan, H., Opachaloemphan, C., Mancini, G., Yang, H., Gallitto, M., Mlejnek, J., Leibholz, A., Haight, K., Ghaninia, M., Huo, L., Perry, M.W., Slone, J., Zhou, X., Traficante, M., Penick, C., Dolezal, D., Gokhale, D., Stevens, D., Fetter-Pruneda, I., Bonasio, R., Zwibel, L.J., Berger, S., Liebig, J., Reinberg, D., and Desplan, C. 2017. An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell, 170: 736-747. PMID: 28802043
- Perry, M.W., Konstantinides, N., Pinto-Teixeira, F., and Desplan, C. 2017. Generation and evolution of neural cell types and circuits: insights from the Drosophila visual system. Annual Reviews of Genetics, 51: 501-527. PMID: 28961025
- Zhang, L., Martin, A., Perry, M.W., Burg, K., Matsuoka, Y., Monteiro, A., and R. Reed. 2017. Genetic basis of melanin pigmentation in butterfly wings. Genetics, 205:1537-1550. PMID: 28193726
- Perry, M.W., Kinoshita, M., Saldi, G., Huo, L., Arikawa, K., and C. Desplan. 2016. Molecular logic behind the three-way stochastic choices that expand butterfly color vision. Nature, 535: 280-284. PMID: 27383790
- Perry, M.W., and C. Desplan. 2016. Quick Guide: Love spots. Current Biology, 26: R1-R2. PMID: 27326705
- Bothma, J.P., Garcia, H.G., Ng, S., Perry, M.W., Gregor, T., and M. Levine 2015. Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. Elife, 4: e07956. PMID: 26267217
- Wernet, M.F, Perry, M.W., and C. Desplan. 2015. The evolutionary diversity of insect retinal mosaics: common design principles and emerging molecular logic. Trends Genetics, 31(6): 316-28. PMID: 26025917
- Perry, M.W., Bothma, J., Luu, R.L., and M. Levine. 2012. Precision of Hunchback expression in the Drosophila embryo. Current Biology, 22: 2247-2252. PMID: 23122844
- Hilgers, V., Perry, M.W., Hendrix, D., Stark, A., Levine, M., and B. Haley. 2011. Neural-specific elongation of 3’ UTRs during Drosophila development. PNAS, 108: 15864-9. PMID: 21896737
- Perry, M.W., Boettiger, A., Levine, M. 2011. Shadow enhancers ensure precision of gap gene expression patterns in the Drosophila embryo. PNAS, 108: 13570-5. PMID: 21825127
- Perry, M.W.*, Boettiger, A.*, Bothma, J. and Levine, M. 2010. Shadow enhancers foster robustness of Drosophila gastrulation. Current Biology, 20: 1562-1567. PMID: 20797865
- Perry, M.W., Boettiger, A., Cande, J. and M. Levine. 2009. Evolution of Insect Dorsoventral Patterning Mechanisms. Cold Spring Harb Symp on Quant Biol, 74: 275-279. PMID: 19843594
- Parchem, R.J., Perry, M.W., and N.H. Patel. 2007. Patterns on the insect wing. Current Opinion in Genetics and Development, 17: 300-308. PMID: 17627807
Biography
Michael Perry got his start in biology while an undergraduate and M.S. student at the University of Florida, working at the Florida Museum of Natural History, where he collected butterflies in the field at sites that ranged from the Rocky Mountains to the cloud forests of Costa Rica. To begin to understand the molecular origins of the diversity he encountered in the field he shifted his focus to developmental biology and gene regulation during his Ph.D. research at UC Berkeley with Mike Levine and Nipam Patel, where he was supported by an NSF-GRFP fellowship. He was able to combine his diverse interests in evolution and developmental biology as a postdoctoral fellow with Claude Desplan at New York University. Michael was the recipient of NIH Ruth L. Kirschstein and Charles H. Revson Foundation fellowships as well as a K99/R00 Pathway to Independence Award from the NIH National Eye Institute. He will join the Division of Biological Sciences faculty at UC San Diego in March 2019.
