Overlooked ‘Housekeeping’ Gene
Plays Unexpected Role in Seizures
Largely ignored molecules expressed in the brain may contribute to imbalances in epilepsy and autism spectrum disorder
September 1, 2020
By Mario Aguilera
Within cells, molecules known as transfer RNAs, or “tRNAs,” play an important but unglamorous workhorse role in keeping the genetic translation process moving along from codes of DNA to functional proteins.
Because they play such a vital role in this translational “housekeeping,” tRNAs are plentiful.
There are hundreds of tRNA genes in mammalian cells and more than enough backup copies, just in case anything goes wrong. Yet because there are so many tRNAs, they’ve been largely overlooked in the search for the roots of disease processes.
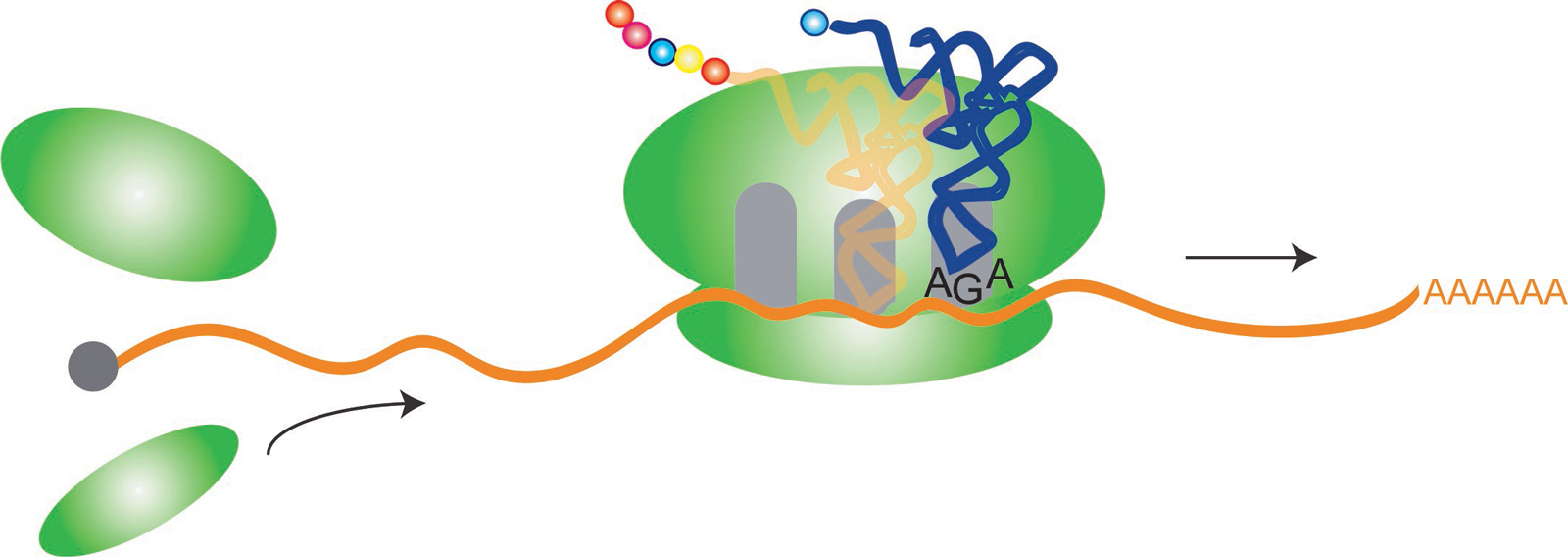
Transfer RNAs, or “tRNAs,” play an important role in keeping the genetic translation process moving along, from codes of DNA to functional proteins.
University of California San Diego scientists studying tRNAs in mice have now found that a mutation in a tRNA gene called n-Tr20—expressed only in the brain—can disrupt the landscape of the entire cell, leading to a chain reaction altering brain function and behavior.
The new research is described August 26 in the journal Neuron.
Study first author Mridu Kapur, a postdoctoral scholar working in Professor Susan Ackerman’s laboratory, says she and her colleagues found that n-Tr20 plays a role in the delicate balance of excitatory and inhibitory neurotransmission in the brain. A disruption in this balance has been implicated in numerous neurological diseases, including epilepsies and autism spectrum disorders.
“tRNAs have generally been overlooked in the hunt for the genetic causes of disease, but recent whole-genome sequencing projects have revealed that there are many variations in tRNA sequences in the human population,” said Kapur. “Our study suggests the enormous potential for tRNA variants to contribute to disease outcomes and phenotypic variability.”
The researchers found that a loss of n-Tr20, one of the members of a five-gene tRNA family, made mice resistant to seizures. While they note that their initial interest in this area came from the idea that a tRNA mutation could subsequently influence other gene mutations, their results not only confirm their speculations that tRNA mutations can influence other mutations, but indicate that these mutations alone can also alter brain function.
“You can imagine it’s like a seesaw—if you push either way you can have problems,” said Ackerman, a member of the Section of Neurobiology, Department of Cellular and Molecular Medicine and investigator at the Howard Hughes Medical Institute. “Keeping balance of these two opposing forces is essential for normal function. Shifting one way or another can lead to neurological diseases. It’s becoming well accepted in the autism spectrum disorder field that what we are really seeing is an imbalance of excitatory/inhibitory neurotransmission.”
Ackerman says part of the reason tRNAs have been overlooked in disease investigations is because researchers commonly concentrate on mutations in unique genes. A member of a large family such as n-Tr20 typically gets tossed in the genetic garbage can because they are too similar to one other.
“We never knew a mutation in a multi-copy tRNA gene could do anything like this,” said Ackerman. “These findings make you think about people who have diseases with variable symptoms and how much this class of overlooked genes could be playing a role in their disease. So we’re seeing this go from a behavior, such as seizure, all the way to the molecular underpinnings causing them.”
The researchers say the results are likely the tip of the iceberg and are now turning their attention to studying tRNA links to disease in tissues outside the brain.
Coauthors of the Neuron paper include Archan Ganguly, Gabor Nagy, Scott Adamson, Jeffrey Chuang and Wayne Frankel.
The research was supported by the National Institutes of Health (R01 NS094637 and R37 NS031349) and the Howard Hughes Medical Institute. Amino acid metabolomics was supported by Mass Spectrometry Core of the Salk Institute (NIH-NCI CCSG P30 014195) and the Helmsley Center for Genomic Medicine.
