Locking Down the Big Bang of Immune Cells
Forgotten strands of DNA initiate the development of immune cells
September 21, 2017
By Mario Aguilera
Intricate human physiological features such as the immune system require exquisite formation and timing to develop properly. Genetic elements must be activated at just the right moment, across vast distances of genomic space.
“Promoter” areas, locations where genes begin to be expressed, must be paired precisely with “enhancer” clusters, where cells mature to a targeted function. Faraway promoters must be brought in proximity with their enhancer counterparts, but how do they come together? When these elements are not in sync, diseases such as leukemia and lymphoma can result. How does this work?
Biologists at the University of California San Diego believe they have the answer.
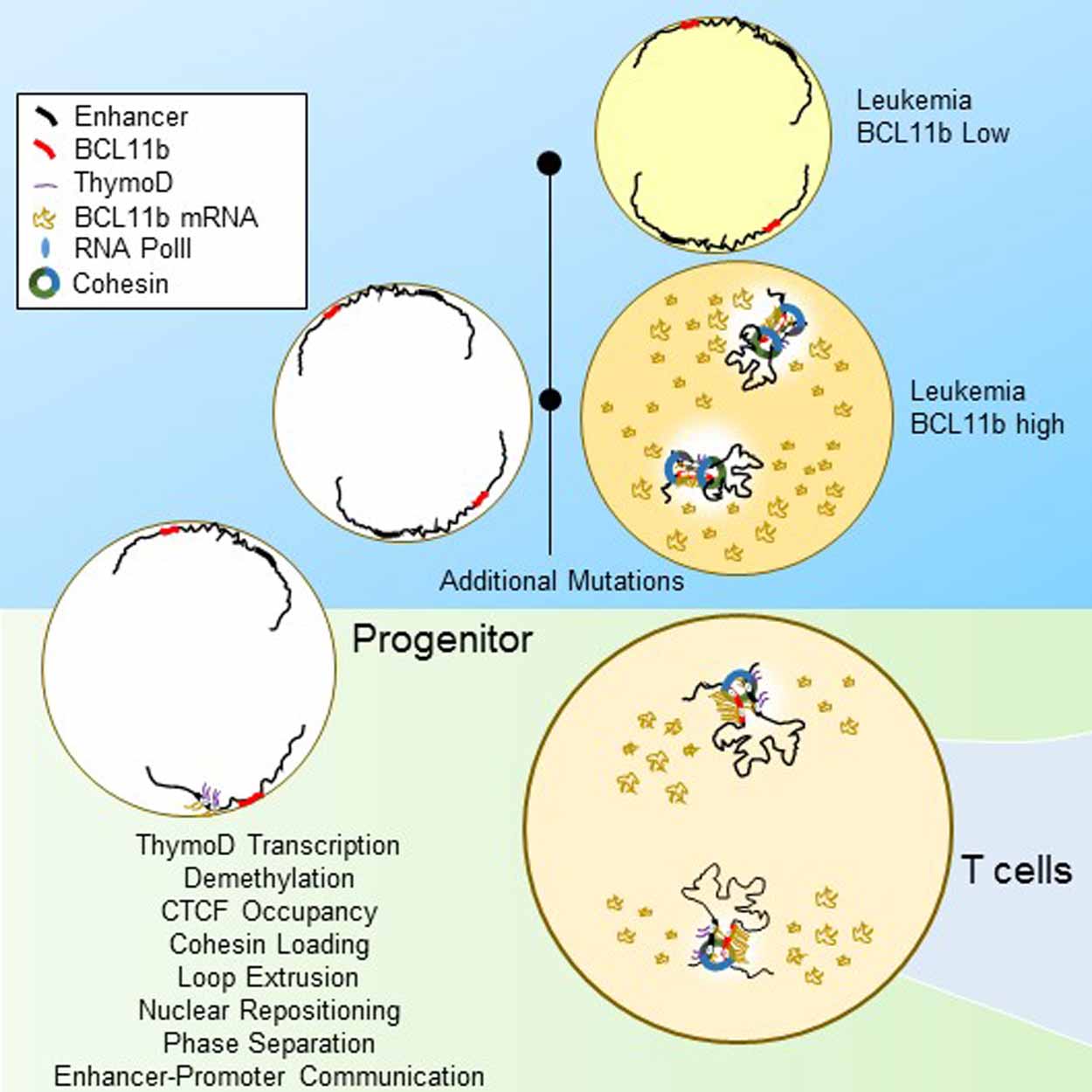
Schematic diagram showing how a subset of immune cells, named DN2a T cells, mature into DN2b T cells. The maturation of this step is among the earliest in immune cell development and is controlled by the forgotten DNA strands that allow the genome to change its architecture to induce the “Big Bang” of T cell development. In the absence of the “forgotten strands” DN2a cells fail to mature and ultimately after accumulating additional mutations become malignant T cells also named leukemias or lymphomas.
Calling it the “big bang” of immune cell development, the researchers made their discovery within previously overlooked stretches of DNA located between genes. The results, published in the September 21 edition of the journal Cell, were led by Takeshi Isoda in Cornelis Murre’s laboratory in UC San Diego’s Division of Biological Sciences.
Through genomic studies and genetic experiments in mice, the scientists found that the ignored areas, known as “non-coding” DNA, activate a change in the 3D structure of DNA that brings promoters and enhancers together with stunning accuracy. Murre describes the mechanism as somewhat like a stiff wire—with enhancers and promoters on either end—that’s bended together into a loop and anchored in place. Enhancers and promoters, once distantly separated, are now repositioned in close proximity to initiate the development of immune system building blocks known as T cells.
“Nature is so clever. We think of the genome as an unstructured strand but in fact what we are seeing is a highly structured and meaningful design,” said Murre. “The process of architecture remodeling we’ve described allows the enhancer and promoter to find each other in 3D space at precisely the right time. The beauty is that it’s all very carefully orchestrated. We have seen one example but there are likely many others all occurring at the same time when cells are moving along the developmental pathway–that’s kind of amazing.”
While Murre and his colleagues concentrated on T cells, they believe this mechanism may be unfolding throughout the animal and plant kingdoms.
When the mechanism fails, T cell development falters and diseases such as lymphoma and leukemia result. Murre says the results show how the forgotten strands of DNA suppress the development of leukemia and lymphoma.
“The implications of these results are not only how normal T cells develop, but that tumor suppression is regulated through this mechanism, at least in part,” said Murre. “Ultimately we may be able to fix mutations associated with disease and these forgotten strands of DNA.”
Coauthors of the paper in include Amanda Moore, Zhaoren He, Vivek Chandra, Masatoshi Aida, Matthew Denholtz, Jan Piet van Hamburg, Kathleen Fisch, Aaron Chang, Shawn Fahl and David Wiest.
The research was supported by the CCBB (Center for Computational Biology and Bioinformatics (UL1TRR001442), the California Institute for Regenerative Medicine (RB5-07025), the National Institutes of Health (AI02853, AI00880 and AI09599) and the Uehara Memorial Foundation.
