Amy Kiger
Research
Cellular remodeling
Investigating membrane regulation of dynamic cell organization & shape
Cells must continuously maintain integrity and compartmentalization with changes in cell size and shape that occur in development, immunity, aging and disease. How cellular spatiotemporal organization is achieved with remodeling is therefore a fundamental problem of broad significance.
Our laboratory is studying central roles for novel membrane trafficking processes, under control of spatiotemporal lipid regulation, that coordinate dynamic cell structure. Using a foundation in genetic and cell biology experiments in the fruit fly, Drosophila, we are linking specific lipid kinase and phosphatase activities to cell remodeling. Our work aims to identify conserved mechanisms of membrane regulation in immune and muscle cell functions, and provide insights into related human diseases.
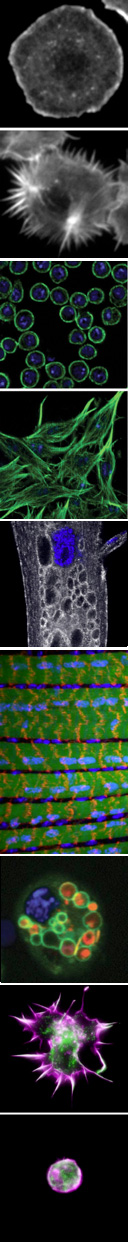
Our research is currently focused in three major areas:
- roles for phosphoinositide lipid regulation in cellular remodeling
- roles for autophagy in cellular remodeling, and
- systems-biology approaches to understanding cellular remodeling
To address these problems, we are investigating questions specific to the dynamic organization of two distinct cell types, immune cells and muscle cells. The different elaborated morphologies and functions of immune cells and muscle cells are helping us to illuminate both broadly conserved and tissue-specific requirements in cellular remodeling.
Regulation of cellular remodeling by phosphoinositide-directed endocytic recycling
Phosphoinositide lipids, the seven different forms of phosphorylated phosphatidylinositol, provide a membrane “code” that controls a wide array of cellular functions. However, gaps remain in understanding the complex functional relationships between the relevant kinases, phosphatases and the specific roles mediated through phosphoinositide regulation. The large family of Myotubularin (MTM) phosphoinositide phosphatases (15 human, 7 fly, 1 yeast) are selective for PI(3)P and PI(3,5)P 2 turnover, pointing to possible roles in complex regulation of endocytosis and/or autophagy. Further confounding the understanding of MTM family functions are that, curiously, half of the members encode for catalytically inactive, or “pseudo”-phosphatases. Importantly, mutations in MTM catalytic and pseudophosphatase members are both associated with human disease, including centronuclear myopathy (XLMTM) and Charcot-Marie-Tooth neuropathy (CMT4B). How potential MTM roles in endocytosis or autophagy relate to disease are not understood.
We identified mtm, a Drosophila MTM1/MTMR2 phosphoinositide phosphatase, as necessary and sufficient for formation of dynamic immune cell protrusions and immune cell recruitment to wounds. We determined that Mtm and Class II PI3-kinase, called Pi3K68D in flies, co-regulates a PI(3)P subpool on endosomes for endocytic recycling important for immune cell dynamic cortical functions. We have also demonstrated a central role for a Pi3K68D/Mtm pathway for phosphoinositide regulation of integrin endocytic trafficking crucial for maintenance of muscle attachments during muscle cell remodeling. We showed that mtm mutant myofibers exhibit hallmarks of the human XLMTM myopathy, and that the fly defects predicted disruption of integrin localization in human XLMTM. The identification of functionally distinct PI(3)P subpools under control of Class II PI3-kinase has broad implications, including therapeutic strategies for MTM-related human disease. Additional implications include possible roles for an Mtm pathway to regulate integrin trafficking in cell migration and metastasis.
In ongoing studies, we are addressing how phosphoinositide-directed endocytic recycling provides key input into the dynamic balance between distinct, cortical F-actin structures in immune cells. In muscle, we are exploring the regulation of integrin trafficking during cellular remodeling, as well as the relationships between mtm and orthologs of other genes also mutated in different forms of human centronuclear myopathy. We are pursuing how PI3KC2 and Mtm are spatiotemporally regulated. Important insights are coming from investigation of the MTM “pseudo”-phosphatase, Sbf, as a key coordinator of both PI3KC2/Mtm phosphoinositide regulation and membrane trafficking.
Regulation of cellular remodeling by autophagy
Autophagy is the membrane-mediated process of cellular “self-eating” through encapsulation of proteins or organelles for lysosomal delivery and degradation, with well-known roles in stress responses. Autophagy has been the focus of growing interest due to its involvement in many processes, including immunity, neurodegeneration and cancer. We showed that autophagy is continuously required for macrophage cell spreading, with functions in proper cytoskeletal reorganization, and has a conserved role in the cortical remodeling of both Drosophila and mouse macrophages. Consequently, macrophages disrupted for autophagy are impaired in their recruitment to epidermal wounds. We showed a requirement for the p62 multiadaptor involved in selective targeting of ubiquitinated proteins for autophagic degradation, pointing to a role for selective autophagy (versus bulk autophagy) as a regulatory mechanism for cortical remodeling. This is the first demonstration of a true “cell remodeling” function – at the level of cell shape – for autophagy, which has been previously proposed but never shown.
In ongoing studies, we are continuing to address how autophagy specifically regulates immune cell remodeling, in part through the use of systems biology approaches to identify targets of selective autophagy. We are also investigating phosphoinositide regulation of autophagy.
Functional genomics of cellular remodeling
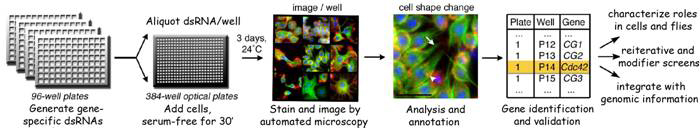
The regulation of cell morphology can be viewed as a complex system that must integrate the temporal-spatial control of multiple cellular processes, including cytoskeletal and membrane dynamics, cell adhesion and growth. To understand morphogenetic programs, we are applying functional genomics to comprehensively probe the cellular-genetic networks that control cellular remodeling. We pioneered RNA-interference (RNAi) screens in Drosophila cells that permit unprecedented genome-wide loss-of-function analysis of metazoan cell morphology. Our RNAi microscopy screens have identified gene functions required to maintain distinct round or flat immune cell morphologies. Now, we are using RNAi screens to study how the addition of a hormonal cue culminates in the change of cells from a round to elongated cell shape, relevant to numerous cellular behaviors in vivo. Furthermore, co-RNAi screens targeting combinations of multiple genes are an ideal method for identifying new components of a pathway through suppressor or enhancer effects. In this way, we have isolated and continue to seek modifiers of specific phosphoinositide and autophagy pathways important in cellular remodeling.
Kiger Lab Select Publications
- Fujita N., W. Huang, T. Lin, J. Groulx, S. Jean, J. Nguyen, Y. Kuchitsu, I. Koyama-Honda, N. Mizushima, M. Fukuda, A. A. Kiger. 2017. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLIFE, 6:e23367. PMID: 28063257.
- Jean S, Cox S, Nassari S, Kiger AA. Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep. (2015) 16:297-311.
- Jean S, Kiger AA. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci. (2014) 127:923-8.
- Jean, S. and A. A. Kiger. 2012. Coordination between Rab GTPase and phosphoinositide regulation and function. Nat Rev Mol Cell Biol13: 463-470.
- Jean, S., S. Cox, E.J. Schmidt, F.L. Robinson and A. Kiger. 2012. Sbf/MTMR13 coordinates PI(3)P and Rab21 regulation in endocytic control of cellular remodeling. Mol Biol Cell23(14):2723-40.
- Ribeiro, I., L. Yuan, G. Tanentzapf, J. Dowling, A. A. Kiger. 2011. Phosphoinositide regulation of integrin trafficking required for muscle attachment and maintenance. PLoS Genet, 7(2): e1001295.
- Velichkova, M., J. Juan, P. Kadandale, S. Jean, I. Ribeiro, V. Raman, C. Stefan and A. A. Kiger. 2010. Drosophila Mtm and class II PI3K coregulate a PI(3)P pool with cortical and endolysosomal functions. J Cell Biol.190(3): 407-25.
- Kadandale, P. and A. A. Kiger. 2010. Role of selective autophagy in cellular remodeling: “self-eating” into shape. Autophagy6:8, 1-2.
- Kadandale, P., J.D. Stender, C.K. Glass and A. A. Kiger. 2010. Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc Natl Acad Sci107(23): 10502-7.
Selected Publications
- Asano Y, Jiménez-Dalmaroni A, Liverpool TB, Marchetti MC, Giomi L, Kiger A, Duke T, Baum B. 2009. Pak3 inhibits local actin filament formation to regulate global cell polarity. HFSP J. 3(3): 194-203.
- Eggert, U. S., A. A. Kiger, C. Richter, Z. E. Perlman, N. Perrimon, T. J. Mitchison and C. M. Field. 2004. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS, 2(12): e379 .
- Boutros*, M., A. A. Kiger*, S. Armknecht, K. Kerr, M. Hild, B. Koch, S. Haas, R. Paro, N. Perrimon . 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science, 303: 832-835.
- Kiger, A. A., B. Baum, S. Jones, M. Jones, A. Coulson, C. Echeverri, N. Perrimon. 2003. A functional genomic analysis of cell morphology using RNA-interference. J. Biol., 2: 27.
- Kiger*, A. A., D. L. Jones*, C. Schulz, M. Rogers and M.T. Fuller. 2001. Stem Cell Self-Renewal Specified by JAK-STAT Activation in Response to a Support Cell Cue. Science 294: 2542-2545 .
- Kiger, A. A., H. White-Cooper and M.T. Fuller. 2000. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature 407: 750-754. *equal authorship.
Biography
Amy Kiger received her Ph.D. as a Howard Hughes Medical Institute Predoctoral Fellow from the Department of Developmental Biology at Stanford School of Medicine. She then completed postdoctoral studies in the Department of Genetics at Harvard Medical School as a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research.As Principal Investigator in her own lab, Amy Kiger has been awarded fellowships from The David & Lucille Packard Foundation, The Sidney Kimmel Foundation for Cancer Research, The March of Dimes and The Hellman Family Foundation, as well as grants from the agencies Human Frontier Science Program and National Institutes of Health.

