Eric Bennett
Research
My lab is interested in understanding the fundamental principles of how proteins are destroyed within cells. The cellular environment must constantly adapt to changing conditions and one mechanistic strategy commonly employed is the targeted modification of proteins by ubiquitin chains and subsequent degradation by the proteasome. The ubiquitin-proteasome system (UPS) can be viewed as the cellular waste management system. The UPS is utilized as the cellular quality control system as it is tasked with the degradation of potentially toxic misfolded and misassembled proteins. The inability of the UPS to properly destroy these proteins has been linked to various human pathologies including amyotrophic lateral sclerosis (ALS, commonly referred to as Lou Gehrig's disease), Parkinson's disease, and many human cancers. Thus, understanding the mechanisms of how the UPS recognizes proteins to be destroyed and how the pathway becomes dysfunctional will be instrumental in understanding the etiology of these diseases. The lab uses integrated approaches to understand UPS activity coupling biochemistry, cell biology, and systems-level approaches.
Deubiquitylating enzyme regulation
The ubiquitylation of substrates occurs via a hierarchy of enzyme activity where ubiquitin activating enzymes (E1s) activate free ubiquitin and transfers the activated ubiquitin to ubiquitin conjugating enzymes, (E2s), who partner with ubiquitin ligases (E3s) to finally conjugate ubiquitin to substrates.
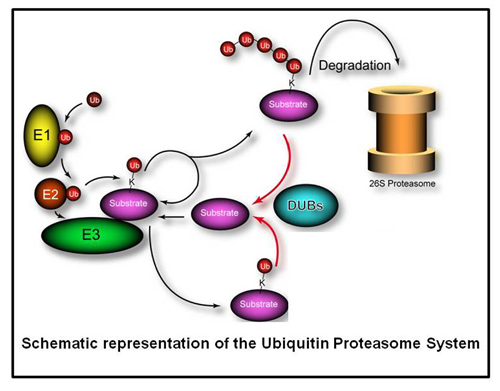
Deubiquitylating enzymes (Dubs) remove ubiquitin chains from substrates thereby antagonizing ubiquitin ligase activity. While the basic enzymatic function of Dubs is understood, how their activity is regulated, what cellular pathways are regulated by Dub activity, and what cellular substrates are the targets for Dub regulationare not fully understood. We previously characterized 75 of the 100 human Dubs based on their cellular interaction partners using proteomic technologies. During this study we established an integrative proteomic platform, termed CompPASS, 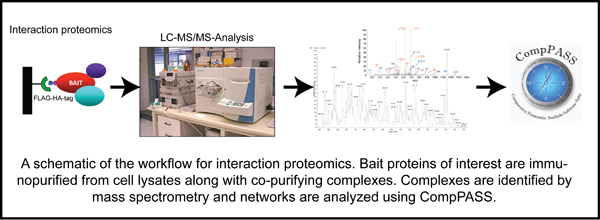
to rapidly analyze interaction proteomics data. We found that Dubs not only frequently associate with ubiquitin ligases but are themselves often modified by ubiquitin. This suggests that Dub activity may be regulated by E3 ligases and vice versa. The lab is interested in understanding the mechanisms of Dub activity regulation via association with ubiquitin ligases using both biochemical and cellular techniques.
Cullin-RING ligase regulation
One family of ubiquitin ligases are the multi-subunit ligases consisting of Cullin-RING ligases (CRLs)
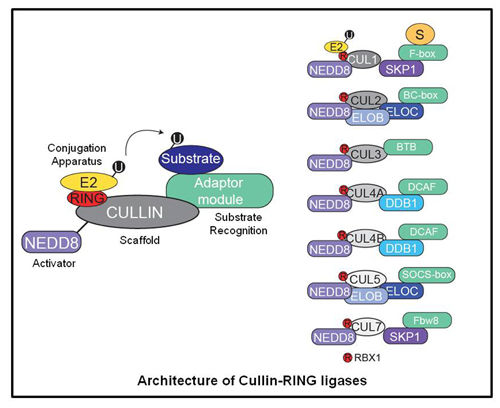
and the anaphase promoting complex (APC). Cullin-RING ligases are modular assemblies that can target many cellular proteins for regulated degradation by dynamically remodeling its subunit architecture. CRLs are known to degrade many critical oncoproteins and tumor suppressors, and as such there has been recent interest in the development of therapeutics targeting this pathway as an anti-cancer strategy.
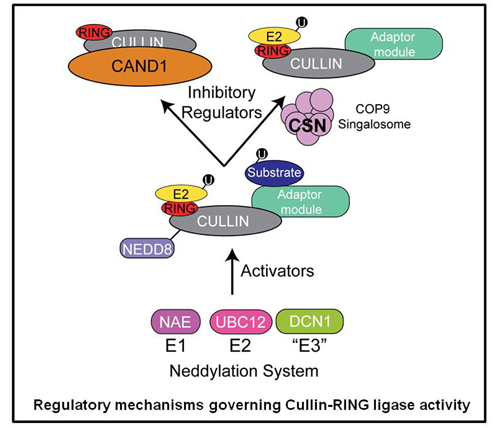
CRLs are also regulated through a switch like mechanism in which the ubiquitin like protein Nedd8 modifies and subsequently activates the cullin subunit. Thus, by simply adding and removing the Nedd8 modification, CRL activity can be switched on and off. We are utilizing quantitative proteomics to study the cellular mechanisms used to control CRL activity. Additionally, this allows us to probe the dynamic nature of CRL assemblies and to understand how their architecture is remodeled to adapt to changes in the cellular environment.
Global quantitative proteomic analysis of UPS function
The cellular proteome is continually monitored by the protein folding and degradation machinery to prevent the accumulation of potentially toxic misfolded or misassembled proteins. Proteins that contain errors in their primary sequence that arise during the translation process need to be recognized and selectively degraded by the UPS. When this quality control function breaks down it can have dire consequences on cellular and organismal fitness. Thus, the ability to globally monitor the activity of the UPS would allow for the identification of conditions that lead to UPS dysfunction. We have developed technologies that allow for the interrogation of the UPS activity on a systematic level. From these studies it is clear that mistranslated proteins are constantly ubiquitylated and cleared from cells. We are interested in trying to answer several questions related to quality control mechanisms. For instance, what are the ubiquitin ligases responsible for ubiquitylation mistranslated substrates? How do changes in free amino acid levels within the cell effect protein degradation and UPS function? Can we monitor alterations in UPS function in human pathologies associated with a decrease in quality control function? How does changing the redox potential within cells effect the quality control machinery? By understanding how proteins are recognized as quality control substrates and how the cell responds to challenges in protein homeostasis we hope to develop therapeutics that modulate quality control function.
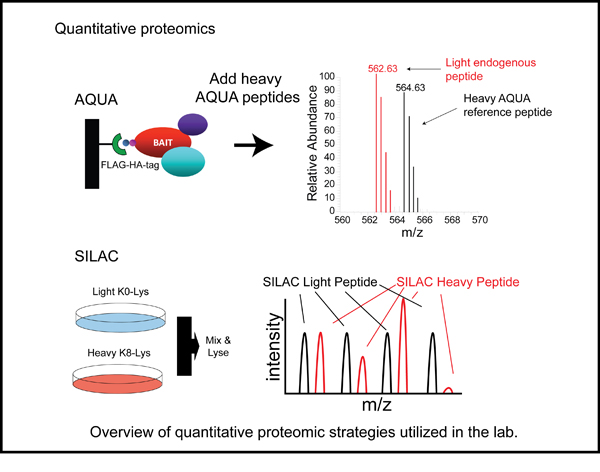
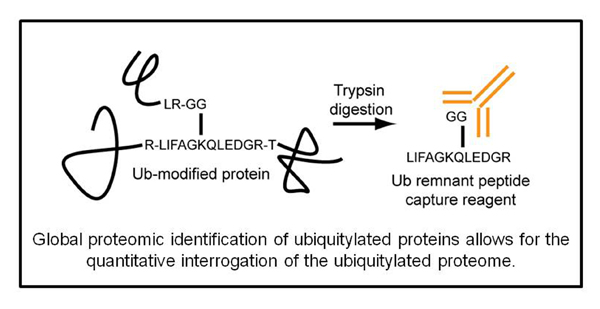
Select Publications
- Sinha NK, Ordureau A, Best K, Saba JA, Zinshteyn B, Sundaramoorthy E, Fulzele A, Garshott DM, Denk T, Thoms M, Paulo JA, Harper W, Bennett EJ, Beckmann R, Green R. EDF1 coordinates cellular responses to ribosome collisions. Elife. 2020 Aug 3;9:e58828. PMID: 32744497
- Xu X, Hou B, Fulzele A, Masubuchi T, Zhao Y, Wu Z, Hu Y, Jiang Y, Ma Y, Wang H, Bennett EJ, Fu G, Hui E. PD-1 and BTLA regulate T cell signaling differentially and only partially through SHP1 and SHP2. J Cell Biol. 2020 Jun 1;219(6):e201905085. PMID: 32437509
- Panek J, Gang SS, Reddy KC, Luallen RJ, Fulzele A, Bennett EJ, Troemel ER. A cullin-RING ubiquitin ligase promotes thermotolerance as part of the intracellular pathogen response in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2020 Apr 7;117(14):7950-7960. PMID: 32193347
- Garshott DM, Sundaramoorthy E, Leonard M, Bennett EJ. Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized. Elife. 2020 Feb 3;9:e54023. PMID: 32011234
- Markmiller S, Fulzele A, Higgins R, Leonard M, Yeo GW, Bennett EJ. Active Protein Neddylation or Ubiquitylation Is Dispensable for Stress Granule Dynamics. Cell Rep. 2019 Apr 30;27(5):1356-1363. PMID: 31042464
- Fulzele A, Bennett EJ. Ubiquitin diGLY Proteomics as an Approach to Identify and Quantify the Ubiquitin-Modified Proteome. Methods Mol Biol. 2018;1844:363-384. PMID: 30242721
- Zuzow N, Ghosh A, Leonard M, Liao J, Yang B, Bennett EJ. Mapping the mammalian ribosome quality control complex interactome using proximity labeling approaches. Mol Biol Cell. 2018 May 15;29(10):1258-1269.PMID: 29540532
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao FB, Bennett EJ, Lécuyer E, Yeo GW. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell. 2018 Jan 25;172(3):590-604. PMID: 29373831
- Lumpkin RJ, Gu H, Zhu Y, Leonard M, Ahmad AS, Clauser KR, Meyer JG, Bennett EJ, Komives EA. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat Commun. 2017 Oct 27;8(1):1171. PMID: 29079793
- Reinke AW, Balla KM, Troemel ER, Bennett EJ. In vivo mapping of tissue- and subcellular-specific proteomes in Caenorhabditis elegans. Sci Adv. 2017 May 10;3(5):e1602426. PMID: 28508060
- Neal S, Mak R, Bennett EJ, Hampton R. A Cdc48 "Retrochaperone" Function Is Required for the Solubility of Retrotranslocated, Integral Membrane Endoplasmic Reticulum-associated Degradation (ERAD-M) Substrates. J Biol Chem. 2017 Feb 24;292(8):3112-3128. PMID: 28077573
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol Cell. 2017 Feb 16;65(4):751-760. PMID: 28132843
- Reinke AW, Balla KM, Bennett EJ, Troemel ER. Identification of microsporidia host-exposed proteins reveals a repertoire of rapidly evolving proteins. Nat Commun. 2017 Jan 9;8:14023. PMID: 28067236
Biography
Dr. Bennett received his PhD from Stanford University. He then carried out postdoctoral work at the Harvard Medical School.

