Jim Wilhelm
Research
Cytoplasmic Organization in Development, Metabolism, and Disease
How is a symmetrical egg turned into an embryo with a head and a tail? How are neuronal connections strengthened or weakened in response to experience? Why don’t incompatible chemical reactions that are necessary for life interfere with each other? The solution to these seemingly unconnected questions lies in the ability of cells to target proteins and nucleic acids to specific subcellular domains. My laboratory is focused on identifying and characterizing novel mechanisms for generating subcellular organization and studying how those mechanisms are used in development, metabolism, and the nervous system. As part of this effort, we use a multidisciplinary approach that applies techniques from biochemistry, genetics, microscopy, and systems biology.
A systems biology approach to identifying novel principles of subcellular organization
In order to identify novel types of intracellular organization, we are conducting a visual screen of the yeast GFP strain collection to identify proteins that assemble into previously unidentified intracellular structures. A pilot version of this screen has identified 38 novel intracellular structures the majority of which are comprised of proteins involved in metabolic pathways – suggesting that self-assembly is a major mechanism for regulating metabolic pathways in vivo.
Consistent with this, one of the proteins we identified, CTP synthase, forms a novel intracellular filament that is conserved from bacteria to mammals. We have also shown that its ability to self-assemble is connected to the regulation of CTP synthase activity. We propose that this form of enzyme regulation functions analogously to the way in which microtubules and actin operate – the core of the polymer is locked into a particular conformation by end limited disassembly. Since we have identified a number of enzymes that exhibit polymerization in vivo, it is likely that this mode of regulation is not unique to CTP synthase.
Interestingly, we have also found that CTP synthase filaments form in axons, but not in dendrites arguing that CTP synthase filaments may also have a cytoskeletal role in neurons apart from their role in regulating enzyme activity. We are currently characterizing CTP synthase filaments in neurons.
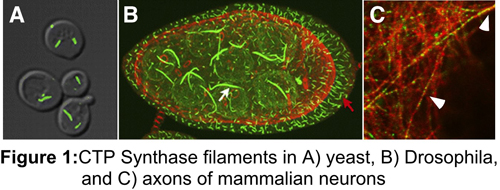
The role of information sorting in Drosophila development
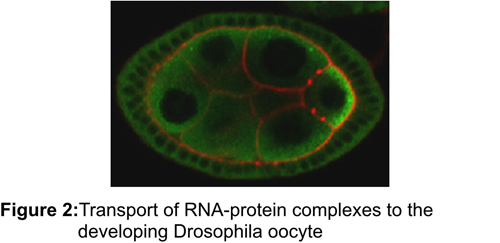
One mechanism for localizing cytoplasmic proteins is to transport the mRNA encoding the protein to the desired location, so that the synthesis of the protein is spatially restricted to particular cytoplasmic regions. Establishment of these domains is further enhanced by a translational control mechanism that ensures that only the properly targeted messages are translated. This sorting mechanism has been implicated in processes as diverse as stem cell differentiation, regulating synaptic strength in neurons and embryonic pattern – underscoring the importance of understanding mRNA localization for medicine, neuroscience, and developmental biology. While the importance of this method of sorting cytoplasmic proteins is clear, it is unclear how localized messages are targeted to different domains or how many cellular or developmental processes utilize mRNA localization. We have used a biochemical approach to purify mRNA transport/translational control complexes from Drosophila melanogaster. By exploiting the powerful genetic tools available in Drosophila, we have begun dissecting how these transport complexes are assembled and disassembled in order to establish the primary body axes of the Drosophila embryo.
Select Publications
- Begovich K, Wilhelm JE. An In Vitro Assembly System Identifies Roles for RNA Nucleation and ATP in Yeast Stress Granule Formation. Mol Cell. 2020 Sep 17;79(6):991-1007.e4. doi: 10.1016/j.molcel.2020.07.017. Epub 2020 Aug 10. PMID: 32780990.
- Begovich K, Vu AQ, Yeo G, Wilhelm JE. Conserved metabolite regulation of stress granule assembly via AdoMet. J Cell Biol. 2020 Aug 3;219(8):e201904141. doi: 10.1083/jcb.201904141. PMID: 32609300; PMCID: PMC7401819.
- Begovich K, Yelon D, Wilhelm JE. PRPS polymerization influences lens fiber organization in zebrafish. Dev Dyn. 2020 Aug;249(8):1018-1031. doi: 10.1002/dvdy.173. Epub 2020 Apr 14. PMID: 32243675; PMCID: PMC8370018.
- Noree C, Begovich K, Samilo D, Broyer R, Monfort E, Wilhelm JE. A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Mol Biol Cell. 2019 Oct 1;30(21):2721-2736. doi: 10.1091/mbc.E19-04-0224. Epub 2019 Sep 4. PMID: 31483745; PMCID: PMC6761767.
- Noree C, Sirinonthanawech N, Wilhelm JE. Saccharomyces cerevisiae ASN1 and ASN2 are asparagine synthetase paralogs that have diverged in their ability to polymerize in response to nutrient stress. Sci Rep. 2019 Jan 22;9(1):278. doi: 10.1038/s41598-018-36719-z. PMID: 30670751; PMCID: PMC6342913.
- Broyer RM, Monfort E, Wilhelm JE. Cup regulates oskar mRNA stability during oogenesis. Dev Biol. 2017 Jan 1;421(1):77-85. doi: 10.1016/j.ydbio.2016.06.040. Epub 2016 Aug 21. PMID: 27554167.
- Noree C, Monfort E, Shiau AK, Wilhelm JE. Common regulatory control of CTP synthase enzyme activity and filament formation. Mol Biol Cell. 2014 Aug 1;25(15):2282-90. doi: 10.1091/mbc.E14-04-0912. Epub 2014 Jun 11. PMID: 24920825; PMCID: PMC4116302.
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010 Aug 23;190(4):541-51. doi: 10.1083/jcb.201003001. Epub 2010 Aug 16. PMID: 20713603; PMCID: PMC2928026.
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007 Mar;175(3):1505-31. doi: 10.1534/genetics.106.065961. Epub 2006 Dec 28. PMID: 17194782; PMCID: PMC1840051.
- Pepling ME, Wilhelm JE, O'Hara AL, Gephardt GW, Spradling AC. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc Natl Acad Sci U S A. 2007 Jan 2;104(1):187-92. doi: 10.1073/pnas.0609923104. Epub 2006 Dec 22. PMID: 17189423; PMCID: PMC1765432.
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006 Dec 21;52(6):997-1009. doi: 10.1016/j.neuron.2006.10.028. PMID: 17178403; PMCID: PMC1955741.
- Wilhelm JE, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell. 2005 Nov;9(5):675-85. doi: 10.1016/j.devcel.2005.09.015. PMID: 16256742.
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol. 2003 Dec 22;163(6):1197-204. doi: 10.1083/jcb.200309088. PMID: 14691132; PMCID: PMC2173729.
- Mansfield JH, Wilhelm JE, Hazelrigg T. Ypsilon Schachtel, a Drosophila Y-box protein, acts antagonistically to Orb in the oskar mRNA localization and translation pathway. Development. 2002 Jan;129(1):197-209. PMID: 11782413.
- Wilhelm JE, Vale RD, Hegde RS. Coordinate control of translation and localization of Vg1 mRNA in Xenopus oocytes. Proc Natl Acad Sci U S A. 2000 Nov 21;97(24):13132-7. doi: 10.1073/pnas.97.24.13132. PMID: 11087864; PMCID: PMC27190.
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000 Oct 13;290(5490):341-4. doi: 10.1126/science.290.5490.341. PMID: 11030653.
- Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J Cell Biol. 2000 Feb 7;148(3):427-40. doi: 10.1083/jcb.148.3.427. PMID: 10662770; PMCID: PMC2174796.
-
Reviews
- Wilhelm, J.E. and Vale, R.D. (1993) RNA on the Move: The mRNA Localization Pathway. J. Cell Biol. 123:269-274.
- Wilhelm, J.E.*, Smibert, C.A.(2005) Mechanisms of translational regulation in Drosophila. Biol Cell. 97(4):235-252.
- * Indicates papers in which Dr. Wilhelm is the corresponding author
Biography
Jim Wilhelm received his Ph.D. in Cell Biology from University of California, San Francisco. He then took a Staff Associate (independent postdoctoral position) in the Department of Embryology, Carnegie Institution of Washington where he received funding from the Life Sciences Research Foundation as a Howard Hughes Medical Institute Fellow. He is also a Sloan Research Fellow in Neuroscience, an Ellison Medical Foundation New Scholar in Aging, and March of Dimes Basil O'Connor Scholar.

