Rick Firtel
Research
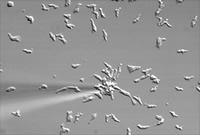
Chemotaxis of wild-type cells to cAMP emitted from a pipette.
Our laboratory focuses on the signal transduction pathways and the mechanics that control directional cell movement, or chemotaxis, and morphogenesis of eukaryotic cells. Chemotaxis is a basic property of many eukaryotic cells and plays critical roles in diverse biological pathways including wound healing, migration of leukocytes to sites of inflammation and bacterial infection, morphogenesis, and metastasis of a variety of cancer cell types. Chemotaxis is mediated by small molecule ligands (chemoattractants and chemokines) that interact with cell surface receptors to control downstream effector pathways. Cells are able to sense and respond to even weak chemoattractant gradients and to amplify a weak chemical gradient into a steep intracellular gradient of signaling molecules. Cells then translate this into changes in the cytoskeleton that control the mechanical forces that drive cell movement.
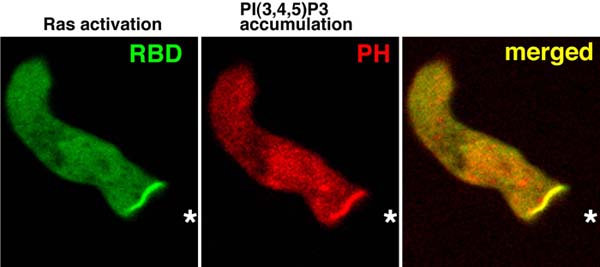
Co-activation of Ras and PI3K at the leading edge of chemotaxing cells using real-time reporters
Over the past few years, great progress has been made in elucidating the molecular mechanisms controlling the ability of cells to sense and respond to chemical gradients. My laboratory and others have discovered that Ras lies at the top of the signal transduction cascade leading to chemotaxis. We demonstrated that Ras controls chemotaxis through the activation of phosphatidylinositol 3-kinase (PI3K) and Tor Complex 2, which are also essential for cell growth and survival in metazoan cells. Our goal is to reveal the molecular mechanisms regulating these and other signaling pathways that enable cells to chemotax. Because most of these components are highly conserved evolutionarily in cells ranging from Dictyostelium amoebae to human leukocytes, we use Dictyostelium as our primary experimental system, which allows us to employ a wide range of genetic as well as biochemical approaches to dissect these signaling pathways. These methods are combined with in vivo, real-time analysis of GFP fusions of signaling proteins in living cells to understand the spatio-temporal changes of these proteins in response to directional signals. These diverse approaches have allowed us elucidate the basic principles by which signal transduction pathways control directional sensing and chemotaxis that apply to human leukocytes as well as Dictyostelium cells.
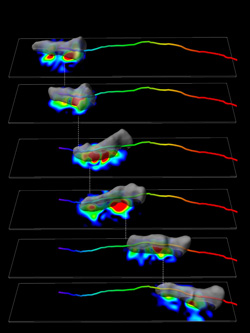
A chemotaxing Dictyostelium cell (gray) forms stationary “traction adhesions” with the substratum along its anterior-posterior axis that transmit force (indicated by heat map) and allow the cell to move. The colored cylinder shows the path of the migrating cell over time.
A major unanswered question in cell motility is how the signaling pathways and the reorganizations of the cytoskeleton function together to control cell movement. In collaboration with the groups of Juan Lasheras and Juan Carlos del Alamo in the Department of Mechanical and Aerospace Engineering, we have developed quantitative approaches to examine the time evolution of the traction forces that mediate each stage of the cell motility cycle. We have made the novel finding that amoeboid cell movement is controlled by a complex interplay between lateral and normal (vertical) forces mediated by cortical tension controlled through the lateral cell cortex as well as the more “classic” F-actin/myosin-mediated axial forces. We have achieved this by integrating the biochemical and mechanical measurements of cell motility and obtained the quantitative information necessary to connect specific biochemical and signaling processes to the physical events regulating the mechanical forces controlling cell motility and, in do so, understand how cells move. In collaboration with Wouter-Jan Rappel, Alex Groisman, and Bill Loomis, we are attempting to mathematically model the signaling events controlling cell polarization.
Select Publications
- Kumagai, A., M. Pupillo, R. Gunderson, R. Miake-Lye, P.N. Devreotes and R.A. Firtel (1989). Regulation and function of G protein subunits in Dictyostelium. Cell 57:265-275. (PMID: 2539262)
- Firtel, R.A. and A.L. Chapman (1990). A role for cAMP-dependent protein kinase in early Dictyostelium development. Genes Devel. 4:18-28.
- Hjorth, A.L., C. Pears, J.G. Williams and R.A. Firtel (1990). A developmentally regulated trans-acting factor(s) recognizes dissimilar G/C-rich elements controlling a class of cAMP-inducible Dictyostelium genes. Genes Devel. 4:419-432.
- Haberstroh, L. and R.A. Firtel (1990). A spatial gradient of expression of a cAMP-regulated prespore cell-type-specific gene in Dictyostelium. Genes Devel. 4:596-612.
- Esch, R.K. and R.A. Firtel (1991). Cyclic AMP and cell sorting control the spatial expression of a developmentally essential cell-type-specific ras gene in Dictyostelium. Genes Devel. 5:9-21.
- Hadwiger, J.A. and R.A. Firtel (1992). Analysis of Gα4, a G protein subunit required for multicellular development in Dictyostelium. Genes Devel. 6:38-49.
- Howard, P., B. Sefton and R.A. Firtel (1992). Analysis of a spatially regulated phosphotyrosine phosphatase identifies tyrosine phosphorylation as a key regulatory pathway in Dictyostelium. Cell 71:637-647.
- Howard, P.K., B.M. Sefton and R.A. Firtel (1993). Tyrosine phosphorylation of actin in Dictyostelium associated with cell shape changes. Science 259:241-244.
- Schnitzler, G.R., W.H. Fischer and R.A. Firtel (1994). Cloning and characterization of the G-box binding factor, an essential component of the developmental switch between early and late development in Dictyostelium. Genes Devel. 8:502-514.
- Dynes, J.L., A.M. Clark, G. Shaulsky, A. Kuspa, W.F. Loomis and R.A. Firtel (1994). LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Devel. 8:948-958.
- Schnitzler, G.R., C. Briscoe, J.M. Brown and R.A. Firtel (1995). Serpentine cAMP receptors may act though a G protein-independent pathway to induce postaggregative development in Dictyostelium. Cell 81:737-745.
- Gaskins, C., A.M. Clark, L. Aubry, J.E. Segall and R.A. Firtel (1996). The Dictyostelium MAP kinase ERK2 regulates multiple, independent developmental pathways. Genes Devel. 10:118-128.
- Ma, H., M. Gamper, C. Parent and R.A. Firtel (1997). The Dictyostelium MAP kinase kinase DdMEK1 regulates chemotaxis and is essential for chemoattractant-mediated activation of guanylyl cyclase. EMBO J. 16:4317-4332.
- Aubry, L and R.A. Firtel (1998). Spalten, a protein containing Gα-protein-like and PP2C domains, is essential for cell-type differentiation in Dictyostelium. Genes Devel. 12:1525-1538.
- Lee, S., C.A. Parent, R. Insall and R.A. Firtel (1999). A novel Ras interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell 10:2829-2845. (PMID: 10473630)
- Chung, C.Y. and R.A. Firtel (1999). PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 147:559-575. (PMID: 10545500)
- Meili, R., C. Ellsworth and R.A. Firtel (2000). A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr. Biol. 10:708-717. (PMID: 10873800)
- Moniakis, J., S. Funamoto, M. Fukuzawa, J. Meisenhelder, T. Araki, T. Abe, R. Meili, T. Hunter, J. Williams and R.A. Firtel (2001). An SH2-domain-containing kinase negatively regulates the phosphatidylinositol-3 kinase pathway. Genes Devel. 15:687-698. (PMID: 11274054)
- Chung, C.Y., G. Potikyan and R.A. Firtel (2001). Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7:937–947. (PMID: 11389841)
- Funamoto, S., K. Milan, R. Meili and R.A. Firtel (2001). Role of phosphatidylinositol 3’ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 153:795-809. (PMID: 11352940)
- Mohanty, S., S. Lee, N. Yadava, M.J. Dealy, R.S. Johnson and R.A. Firtel (2001). Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Devel. 15:1435-1448. (PMID: 11390363)
- Sobko, A., H. Ma, and R A. Firtel (2002). Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Devel. Cell 2:745-756. (PMID: 12062087)
- Funamoto, S., R. Meili, S. Lee, L. Parry and R.A. Firtel (2002). Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109:611-623. (PMID: 12062104)
- Sasaki, A.T., C. Chun, K. Takeda and R.A. Firtel (2004). Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167:505-518. (PMID: 15534002)
- Park, K.C., F. Rivero, R. Meili, S. Lee, F. Apone and R.A. Firtel (2004). Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 23:4177-4189. (PMID: 15470506)
- Lee, S., F.I. Comer, A. Sasaki, I.X. McLeod, Y. Duong, K. Okamura, J.R. Yates III, C.A. Parent and R.A. Firtel (2005). TOR Complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell 16:4572-4583. (PMID: 16079174)
- Jeon, T.J., D. Lee, S. Merlot, G. Weeks and R.A. Firtel (2007). Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J. Cell Biol. 176:1021-1033. (PMID: 17371831)
- Sasaki, A., C. Janetopoulos, S. Lee, P.G. Charest, K. Takeda, L.W. Sundheimer, R. Meili, P.N. Devreotes and R.A. Firtel (2007). G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 178:185-191. (PMID: 17635933)
- Del Alamo, J.C., R. Meili, B. Alonso-Latorre, J. Rodriguez-Rodriguez, A. Aliseda, R.A. Firtel and J.C. Lasheras (2007). Spatio-temporal analysis of eucaryotic cell motility by improved force cytometry. Proc. Natl. Acad. Sci. USA 104:13343-13348. (PMID: 17684097)
- Jeon, T.J., D. Lee, S. Lee, G. Weeks and R.A. Firtel (2007). Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotaxing cells. J. Cell Biol. 179:833-843. (PMID: 18039932)
- Zhang, S., P.G. Charest and R.A. Firtel (2008). Spatiotemporal regulation of Ras activity provides directional sensing. Curr. Biol. 18:1587-1593. (PMID: 18948008)
- Meili, R., B. Alonso-Latorre, J.C. del Alamo, R.A. Firtel and J.C. Lasheras (2010). Myosin II is essential for the spatiotemporal organization of traction forces during cell motility. Mol. Biol. Cell 21:405-417. (PMID: 19955212)
- Charest, P.G., Z. Shen, A. Lakoduc, A.T. Sasaki, S. Briggs and R.A. Firtel (2010). A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Devel. Cell 18:737-749. (PMID: 2049388)
- Lee, S., Z. Shen, D.N. Robinson, S. Briggs and R.A. Firtel (2010). Involvement of the cytoskeleton in controlling leading-edge function during chemotaxis. Mol. Biol. Cell 21:1810-1824. (PMID: 20375144)
- Bastounis, E., R. Meili, B. Alonso-Latorre, J.C. del Alamo, J.C. Lasheras and R.A. Firtel (2011). The Scar/WAVE complex is necessary for proper regulation of traction stresses during amoeboid motility. Mol. Biol. Cell 22:3995-4003. (PMID: 21900496)
- Takeda, K., D. Shao, M. Adler, P.G. Charest, W.F. Loomis, H. Levine, A. Groisman, W. Rappel and R.A. Firtel (2012). Incoherent feedforward control governs adaptation of activated Ras in a eukaryotic chemotaxis pathway. Science Signaling 5:ra2. (PMID: 22215733)
- Bastounis, E., R. Meili, B. Alvarez-Gonzalez, J. Francois, J.C. del Alamo, R.A. Firtel and J.C. Lasheras (2014). Both contractile axial and lateral traction force dynamics drive amoeboid cell motility. J. Cell Biol. 204:1045-1061. (PMID: 24637328)
- Álvarez-González B, R. Meili, E. Bastounis, R.A. Firtel, L.C. Lasheras, J.C. Del Álamo. (2015). Three-dimensional balance of cortical tension and axial contractility enables fast amoeboid migration. Biophys J. 108:821-832. (PMID: 25692587)
Biography
Rick Firtel was an undergraduate in Biology and Chemistry and Rufus Choate Scholar at Dartmouth College (1966) and received his PhD at Caltech in James Bonner’s laboratory as an NSF fellow (1971). Rick did his postdoctoral work with Harvey Lodish at MIT as a Helen Hay Whitney Foundation Fellow and joined the faculty at UCSD in 1973. He has been the recipient of an American Cancer Society Faculty Research Award, an International Union Against Cancer Fellowship, a Japanese Society for the Promotion of Sciences Fellowship, and a John Simon Guggenheim Fellowship. He has served as Director of the Center for Molecular Genetics (1995-2004), Chair of the Section of Cell and Developmental Biology (1999-2006), and Associate Dean of Biological Sciences (2008-2017).

