Christopher Lee
Research
The goal of the lab is to develop multiscale physical models which bring to life biological scenes at the subcellular and cellular scales--just as molecular dynamics has brought to life static crystal structures of proteins and found varied applications in drug discovery. We develop new modeling approaches which can complement clever experimental control of model organisms/systems to derive new biophysical insights. Returning to the protein dynamics analogy, the familiar relationship between sequence ↔ structure/dynamics ↔ function for proteins, has a counterpart at cellular and subcellular scales: chemically detailed membrane-protein interactions generate forces to deform membranes leading to familiar cell shapes (e.g., the folded cristae of mitochondria) which are often central to biological function. In spite of this general dogma, and despite the numerous correlations between cell shape and disease across a wide variety of contexts, the details behind the biophysics governing these phenomena are unknown. Predicting/discovering the causal mechanisms behind cell shape and disease are our key scientific goals which drive our research direction and modeling innovations.
Select Publications
- F. Yuan, C. T. Lee, J. Houser, A. Sangani, L. Wang, E. Lafer, P. Rangamani#, and J. Stachowiak. “The ins and outs of membrane bending by intrinsically disordered proteins”. Science Advances 9.27 (July 7, 2023), eadg3485. DOI: 10.1126/sciadv.adg3485. PMC: 10328403.
- H. Nakamura#, E. Rho, C. T. Lee, K. Itoh, D. Deng, S. Watanabe, S. Razavi, H. T. Matsubayashi, C. Zhu, E. Jung, P. Rangamani, S. Watanabe, and T. Inoue#. “ActuAtor, a Listeria-inspired molecular tool for generating force in living cells: Controlled deformation of intracellular organizations”. Cell Reports 42.10 (Oct. 2023), p. 113089. DOI: 10.1016/j.celrep.2023.113089. PMC: 10872831.
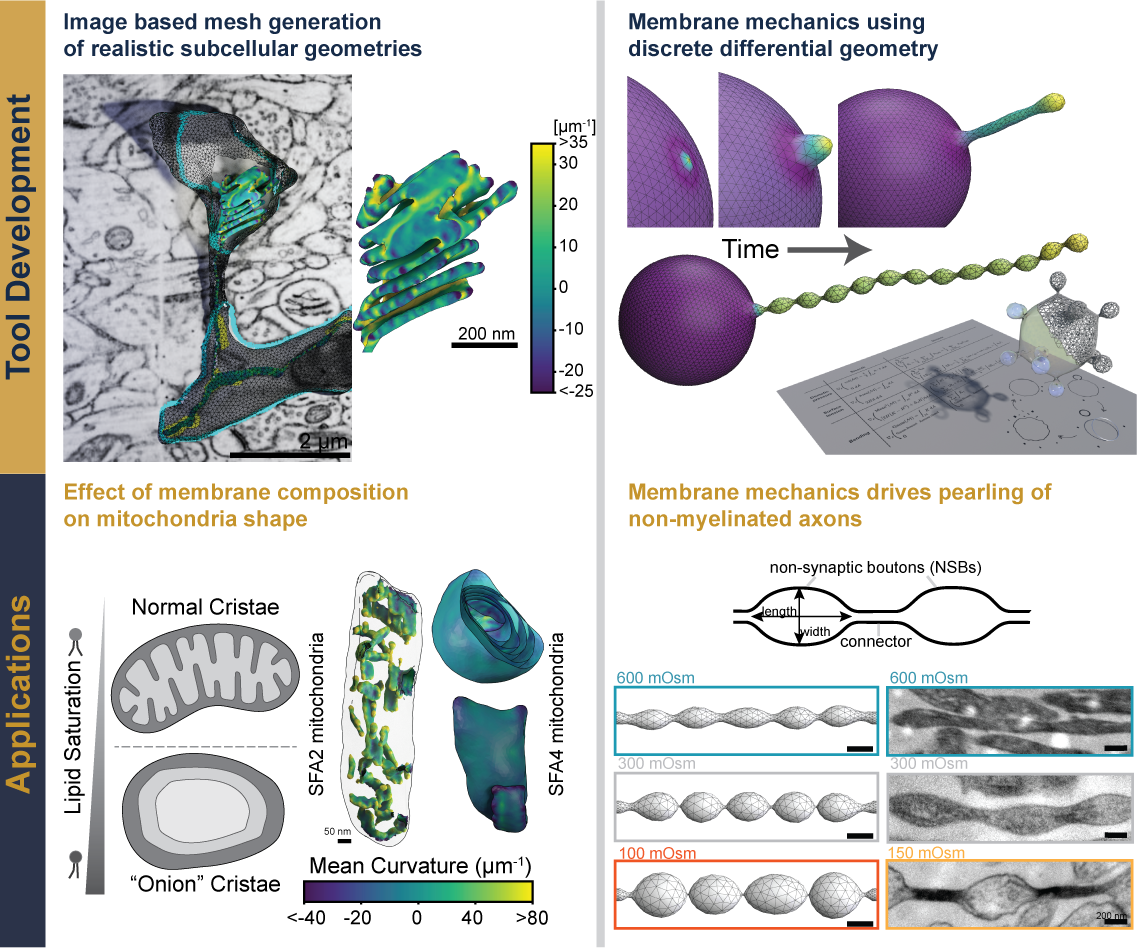
- K. Venkatraman, C. T. Lee†, G. C. Garcia†, A. Mahapatra†, G. Perkins, K.-Y. Kim, H. A. Pasolli, S. Phan, J. Lippincott-Schwartz, M. Ellisman, P. Rangamani#, and I. Budin#. “Cristae formation is a mechanical buckling event controlled by the inner membrane lipidome”. The EMBO Journal 42.24 (Dec. 11, 2023), e114054. DOI: 10.15252/embj.2023114054. PMC: 10054968.
- M. K. Bell†, M. V. Holst†, C. T. Lee, and P. Rangamani#. “Dendritic Spine Morphology Regulates Calcium-Dependent Synaptic Weight Change”. J. Gen. Physiol. 154.8 (July 1, 2022), e202112980. DOI: 10.1085/jgp.202112980. PMC: 9280073.
- C. Zhu, C. T. Lee#, and P. Rangamani#. “Mem3DG: An open-source software framework for 3-D membrane mechanochemical dynamics using discrete differential geometry on triangulated meshes”. Biophys. Reports 2.3 (Sept. 14, 2022), p. 100062. DOI: 10.1016/j.bpr.2022.100062. PMC: 9495267.
- C. T. Lee†, J. G. Laughlin†, N. Angliviel de La Beaumelle, R. E. Amaro, J. A. McCammon, R. Ramamoorthi, M. J. Holst, and P. Rangamani#. “3D Mesh Processing Using GAMer 2 to Enable Reaction-Diffusion Simulations in Realistic Cellular Geometries”. PLOS Comp. Biol. 16.4 (Apr. 6, 2020), e1007756. DOI: 10.1371/journal.pcbi.1007756. PMC: 6716611.
† equal contribution; # corresponding
Biography
Chris Lee received a B.Sc. in chemistry, B.A. in computer science and a M.Sc. in biochemistry from the University of Virginia. He received his Ph.D. in 2019 from UC San Diego, where he worked with Drs. Rommie Amaro and J. Andrew McCammon on molecular simulations. He continued his research at UC San Diego working with Drs. Padmini Rangamani and Michael Holst working on models for cellular membrane mechanics and cell signaling. He joined the Molecular Biology faculty at UC San Diego in 2024.