Eric Allen
Research
Research in our laboratory centers upon the use of environmentally-derived genome sequence information (“metagenomics”) to explore the genetic potential, ecology, and evolution of environmental microbial populations. The nature of this work relies equally upon field-based collections, bioinformatics (tool development, genome assembly, annotation, and comparative analyses) and the tools of molecular ecology and genetics. Together, these approaches enable us to utilize environmental genome sequence data to understand natural microbial phenomena including metabolic activities, biosynthetic processes, environmental adaptation, lateral gene transfer events, biogeographical patterning, biogeochemical cycling, and microbe-microbe/microbe-host interactions. Current projects include:
Marine Metazoan Microbiomes
Marine microorganisms are major contributors to biogeochemical cycling and the maintenance of global homeostasis, yet relatively little is known about the diversity and function of microbes found in intimate association with marine animals. Our current understanding of these complex microbial communities has largely been built upon cultivation-based studies and limited contemporary studies using cultivation-independent methods that survey 16S rRNA gene marker diversity. We use both vertebrate (marine fish) and invertebrate (sponge, mollusks) to investigate the host-associated microbial biology of our oceans, employing metabolic functional gene markers and deep metagenomic sequencing and assembly to complement additional phylogenetic techniques such as 16S rRNA gene amplicon sequencing. We are also developing laboratory-based bioreactor cultivation systems to propagate host-associated microbial communities under controlled conditions to allow experimental manipulation of host microbiota in vitro. These studies are revealing novel microbial diversity previously unidentified in the marine environment and provide new insight into symbiotic relationships between marine animals and their microbial partners.
Genomics of Marine Microbial HOC Synthesis
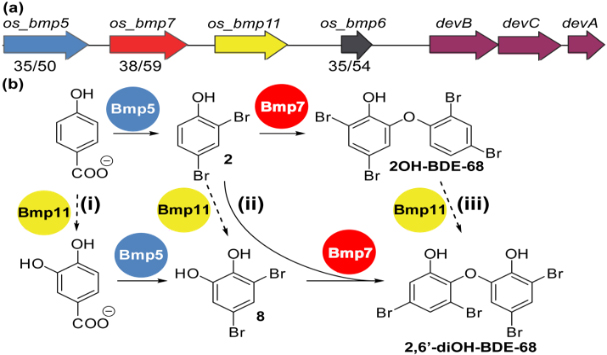
Halogenated organic compounds (HOCs) are widely distributed among organisms occupying higher trophic levels of the marine food web. Chemical analysis of environmental and biological samples has shown that these HOCs originate from both natural and anthropogenic sources. In this work we seek a molecular-level understanding of the microbial genes and enzymatic processes responsible for the biosynthesis of HOCs by marine microorganisms, with the ultimate goal of enabling genetic source tracking through DNA sequence-based biosynthetic signatures, across all trophic levels of the marine food web. In addition to the biosynthesis of these compounds, we are also investigating microbial activities that degrade or transform these pollutants within marine animals, e.g. the fish gut microbiome.
Omega-3 Polyunsaturated Fatty Acids
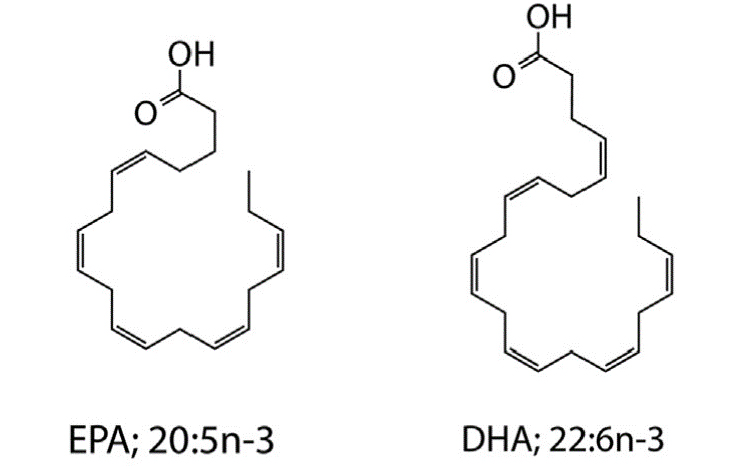
Long-chain omega-3 polyunsaturated fatty acids (PUFAs) such as eicosapentaenoic acid (EPA; 20:5n-3) and docosahexaenoic acid (DHA; 22:6n-3) are well known for their beneficial roles in human health and development, and their ameliorative activities in diverse disease conditions, including cardiovascular, inflammatory, autoimmune, and neurodegenerative disorders. Select lineages of cosmopolitan marine prokaryotic and eukaryotic microorganisms synthesize these compounds via a unique fatty acid synthase/polyketide synthase mechanism that is distinct from the canonical desaturase/elongase-mediated pathway employed by the majority of eukaryotic single-cell microorganisms and metazoans. We are investigating the genetics and regulation of bacterial omega-3 PUFA synthesis in several lineages of marine bacteria via bacterial genetic methods to uncover genetic determinants involved in PUFA synthesis and explore the physiological role of these unique marine lipids. In this process we have engineered native and recombinant bacterial strains that synthesize high quantities of these molecules, providing a novel route for the biotechnological and/or nutraceutical exploitation of these marine bacterial resources.
Bioinformatics
We develop and apply bioinformatics tools and pipelines for the annotation and analysis of microbial genomes and metagenomes, collaborating with colleagues in many diverse areas of experimental biology and microbial ecology, as well as marine natural product chemistry and marine geochemistry. Our special interests include metagenomic sequence assembly of environmental and host-associated DNA samples, determining the role of specific environmental habitats in cross-species transmission of microbial genomes via horizontal gene transfer, and developing in silico tools for the discovery and novelty assignment of secondary metabolite biosynthesis genes within microbial genomes and metagenomes.
Select Publications
- Podell, S, Blanton JM, Neu A, Agarwal V, Biggs JS, Moore BS, Allen EE. 2019. Pangenomic comparison of globally distributed Poribacteria associated with sponge hosts and marine particles. ISME Journal 13:468-481. DOI: 10.1038/s41396-018-0292-9
- Allemann, MN, Shulse CN, Allen EE. 2019. Linkage of marine bacterial polyunsaturated fatty acid and long-chain hydrocarbon biosynthesis. Front Microbiol 10:702. DOI: 10.3389/fmicb.2019.00702
- Schorn, MA, Jordan PA, Podell S, Blanton JM, Agarwal V, Biggs JS, Allen EE, Moore BS. 2019. Comparative genomics of cyanobacterial symbionts reveals distinct, specialized metabolism in tropical Dysideidae sponges. mBio 10:e00821-19. DOI: 10.1128/mBio.00821-19
- Neu, AT, Allen EE, Roy K. 2019. Diversity and composition of intertidal gastropod microbiomes across a major marine biogeographic boundary. Environ Microbiol Rep 11:434-447. DOI: 0.1111/1758-2229.12743
- Kolody, B, McCrow J, Zeigler Allen L, Aylward F, Fontanez K, Moustafa A, Moniruzzaman M, Chavez F, Scholin C, Allen EE, Worden A, DeLong EF, Allen AE. 2019. Diel transcriptional response of a California Current plankton microbiome to light, low iron, and enduring viral infection. ISME Journal (in press)
- Allemann, MN, Allen EE. 2018. Characterization and application of marine microbial omega-3 polyunsaturated fatty acid synthesis. Methods in Enzymology 605:3-32. DOI: 10.1016/bs.mie.2018.02.018
- Minich, JJ, Humphrey G, Benitez RAS, Sanders J, Swafford A, Allen EE, Knight R. 2018. High-throughput miniaturized 16S rRNA amplicon library preparation reduces costs while preserving microbiome integrity. mSystems 3:e00166-18. DOI: 10.1128/mSystems.00166-18
- Agarwal, V, Blanton JM, Podell S, Taton A, Schorn MA, Busch J, Lin Z, Schmidt EW, Jensen PR, Biggs JS, Golden JW, Allen EE, Moore BS. 2017. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nature Chemical Biology 13:537-543. DOI: 10.1038/nchembio.2330
- Agarwal, V, Gamal AEA, Yamanaka K, Poth D, Kersten RD, Schorn M, Allen EE, Moore BS. 2014. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nature Chemical Biology 10:640-647. DOI: 10.1038/nchembio.1564
- Podell, S, Emerson JB, Jones CM, Ugalde JA, Welch S, Heidelberg KB, Banfield JF, Allen EE. 2014. Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community. ISME Journal 8:979-990. DOI: 10.1038/ismej.2013.221
Biography
Eric Allen received his B.S. from the University of Oregon and his Ph.D. from the Scripps Institution of Oceanography at UC San Diego. He performed postdoctoral research at the University of California, Berkeley as an NSF Postdoctoral Fellow in Microbial Biology and joined the faculty of Biological Sciences and the Scripps Institution of Oceangraphy at UCSD in 2006.

