James T. Kadonaga
Research
Transcriptional Regulation and Chromatin Dynamics
Many biological phenomena, including numerous human diseases, can be traced to the proper or improper expression of a gene or set of genes. Hence, the understanding of the mechanisms of gene regulation is essential for the successful analysis of a wide range of problems in the biological and biomedical sciences.
In my lab, we study the regulation of transcription by RNA polymerase II. A surprising number of fundamental aspects of this critically important process have yet to be discovered and characterized. Fortunately, we are now in a new and exciting era in the analysis of gene regulation, as recent and emerging technologies are enabling the illumination of previously unattainable insights into key unanswered questions.
We are also interested in the biological functions of chromatin, the natural state of DNA in the eukaryotic nucleus. Chromatin is complex, multidimensional, and intellectually fascinating. Ultimately, DNA-directed processes in the nucleus must be understood in the context of chromatin.
Some of our research is summarized as follows.
The RNA Polymerase II Core Promoter
In the area of transcription, we have focused on the RNA polymerase II core promoter. The core promoter is the stretch of DNA that directs transcription initiation, and it typically comprises the region from about -40 to +40 nt relative to the +1 transcription start site. The core promoter is sometimes called the "gateway to transcription", as it is the site of convergence of the signals that activate transcription.
Core promoters are diverse in terms of their structure and function. The transcriptional activity of core promoters is driven by DNA elements such as the TATA box, initiator (Inr), TCT motif, downstream core promoter element (DPE), motif ten element (MTE), and others (Fig. 1). There are no universal core promoter elements. Any particular core promoter might have one, some, or none of these DNA elements. In addition, there are core promoter elements that remain to be discovered.
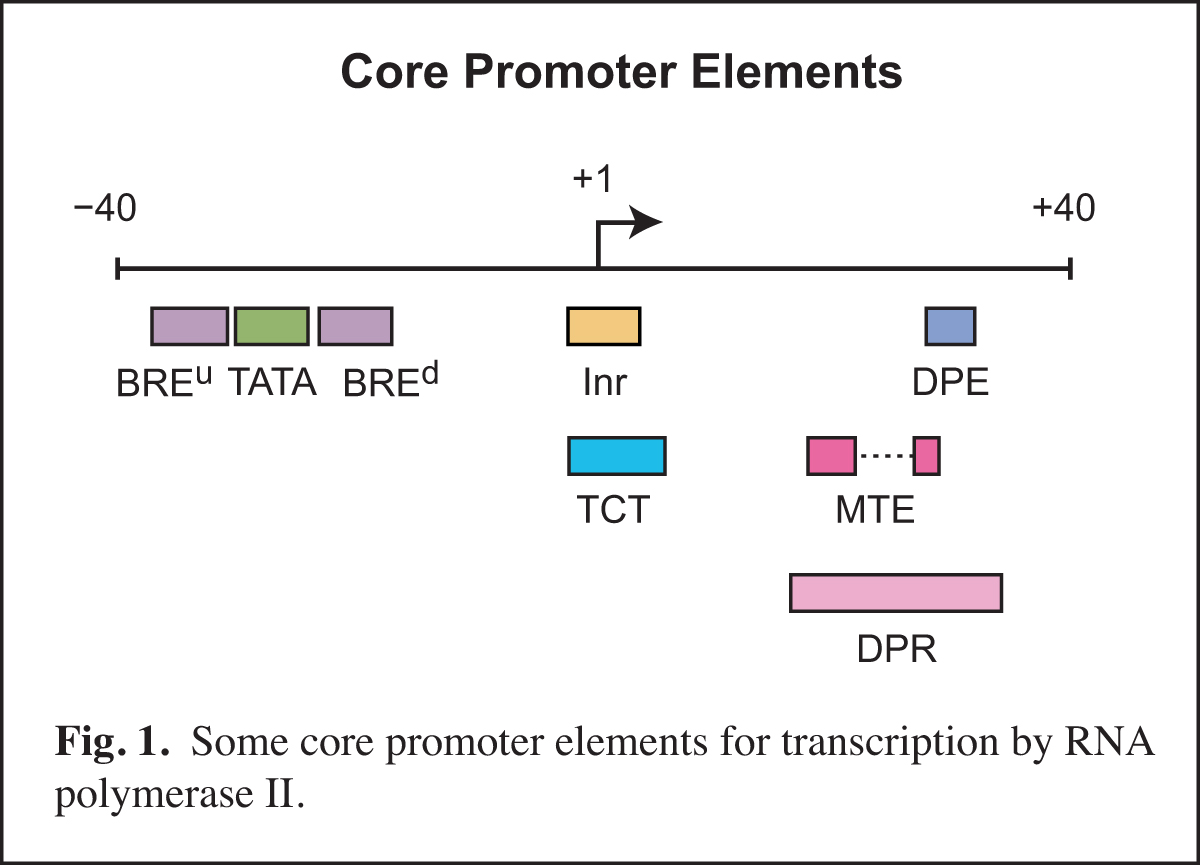
We study core promoters at three levels: DNA elements, transcription factors, and biological networks.
(1). RNA polymerase II core promoter: DNA elements
First, it is essential to identify the DNA-based signals that direct the initiation of transcription. In some cases, these signals can be seen as DNA sequence signatures, such as a TATAAA-like sequence for the TATA box. As described below, however, the DNA sequence signatures of transcriptional elements are not always clearly evident.
In Drosophila, we identified and characterized the DPE core promoter element and also articulated the nature and properties of the MTE and TCT motifs (for recent reviews, see: Vo ngoc, Wang, et al., 2017; Vo ngoc et al., 2019).
More recently, we have been studying the human core promoter, which is much more abstruse than the Drosophila core promoter. As a first step in this endeavor, we generated and analyzed genome-wide data on transcription start sites (TSSs) and identified the DNA sequence signature of the human initiator (Inr) element (Vo ngoc, Cassidy, et al., 2017).
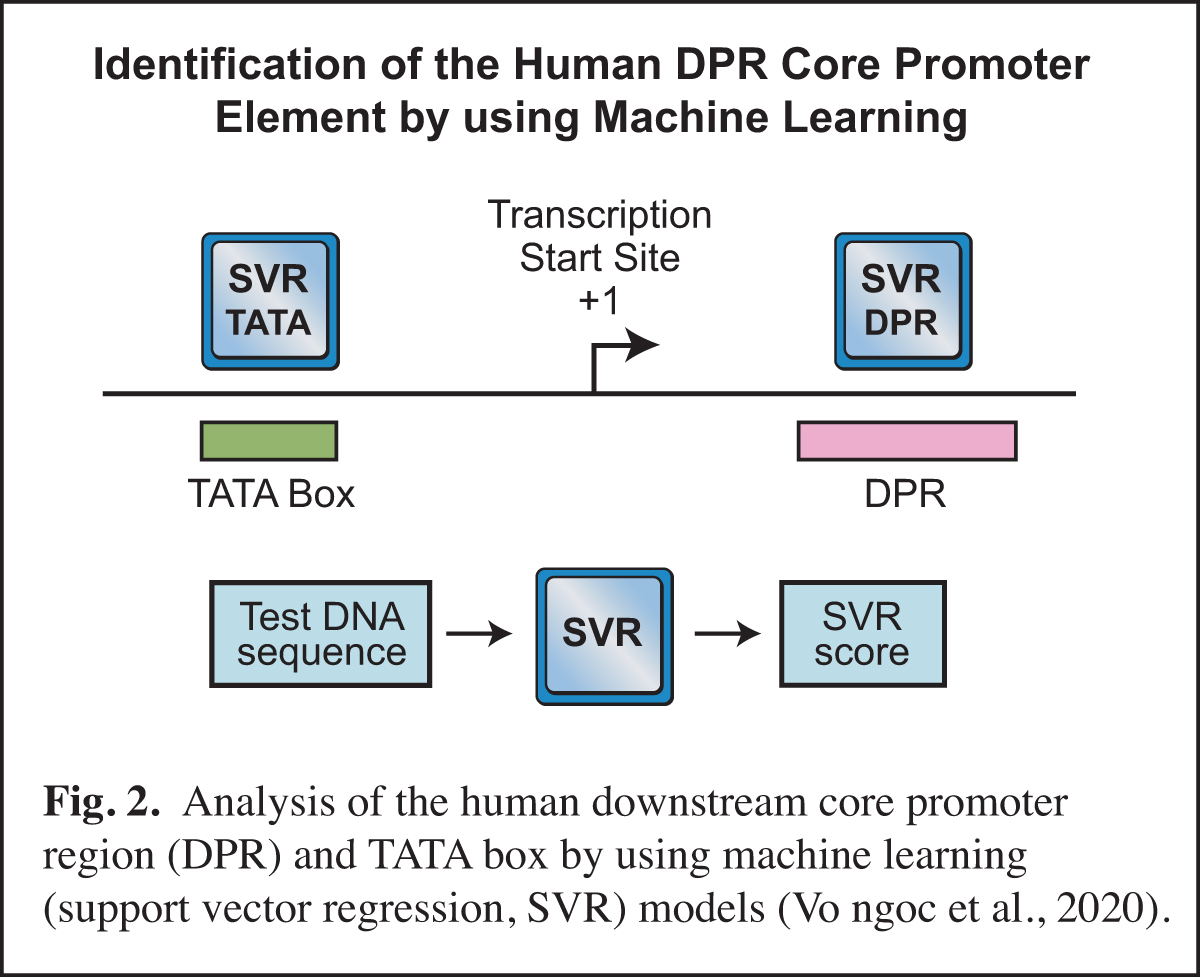
In humans, the analysis of the downstream core promoter region was particularly challenging. In Drosophila, the downstream MTE and DPE elements are commonly found and have been extensively characterized, but it was difficult to discern whether or not these elements exist in humans. This puzzle was ultimately solved by using machine learning (Vo ngoc et al., 2020) (Fig. 2). In this work, we generated a machine learning (support vector regression, SVR) model for the human downstream core promoter region (DPR), which encompasses the positions of the Drosophila MTE and DPE (Fig. 1). The SVR model was accurately able to predict the presence of an active DPR element in humans.
The human DPR is too complex to be described by a DNA sequence signature, but its essential characteristics could be deciphered by machine learning. We also generated an SVR model for the human TATA box. Importantly, these SVR models provide a quantitative prediction of DPR or TATA box activity. For many reasons (discussed in Vo ngoc et al., 2020), the SVR models are much more effective than DNA sequence signatures for predicting the presence and activities of the DPR and TATA elements. These studies have opened up exciting new avenues in our analysis of the core promoter.
(2). RNA polymerase II core promoter: Transcription factors
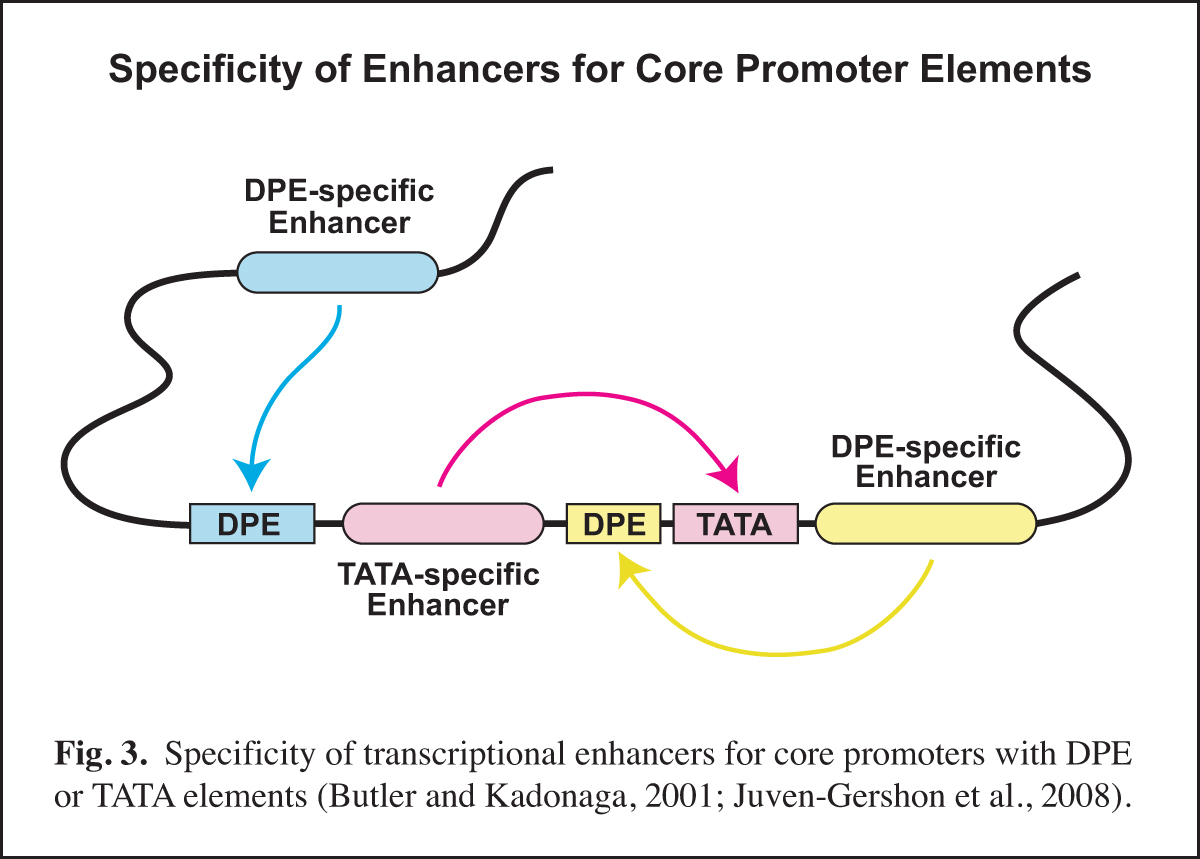
Next, it is important to identify and to characterize the transcription factors that function with the specific core promoter elements. In this regard, it is important to remember that the core promoter is a diverse regulatory element. Different types of core promoters (for example, TATA-dependent versus DPE-dependent promoters) are transcribed by different mechanisms and can be differentially regulated by transcriptional enhancers.
In Drosophila, we found that TATA-less transcription with TCT-dependent core promoters does not require TBP (TATA box-binding protein), but is instead dependent upon TRF2 (TBP-related factor 2). We also discovered that Caudal protein (a DNA-binding transcription factor that is a key regulator of the Hox genes) is a DPE-specific activator. This finding is a specific example of the concept that some transcriptional enhancers are specific for core promoters with either a DPE or a TATA element (Fig. 3). With the identification of the human DPR, it is now possible to investigate factors that differentiate between TATA and DPR elements in humans.
(3). RNA polymerase II core promoter: Biological networks
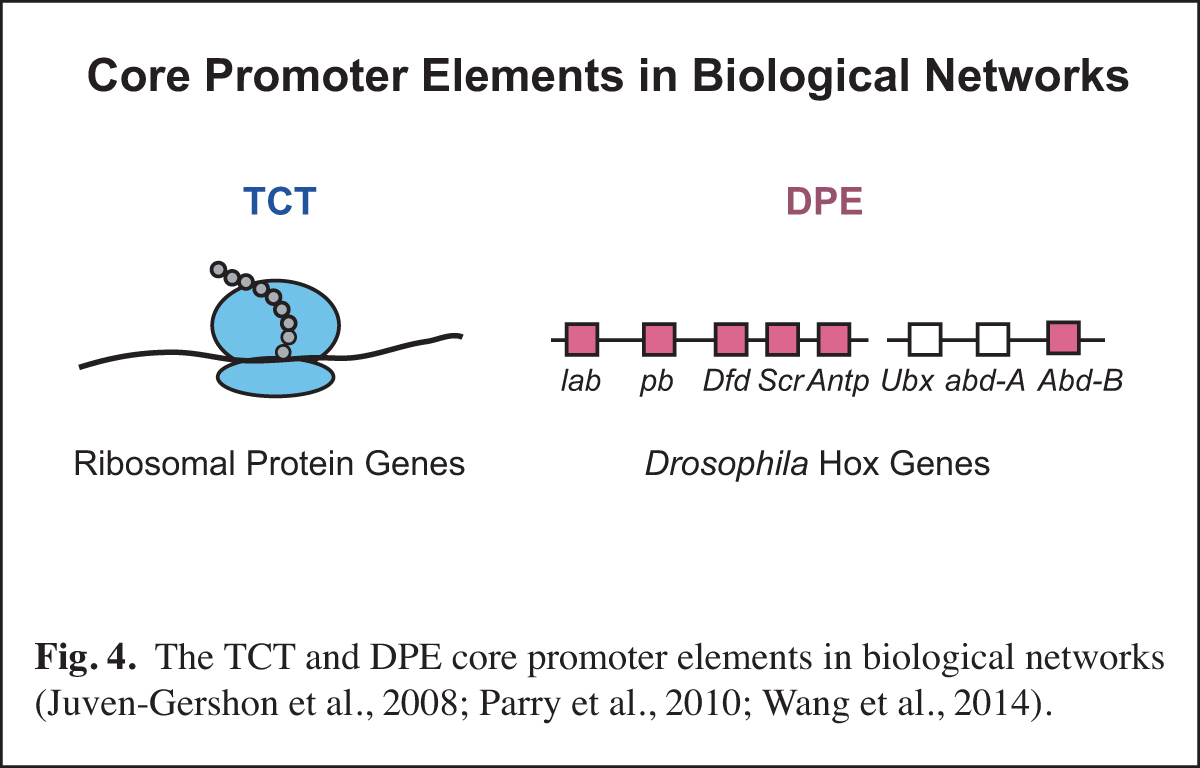
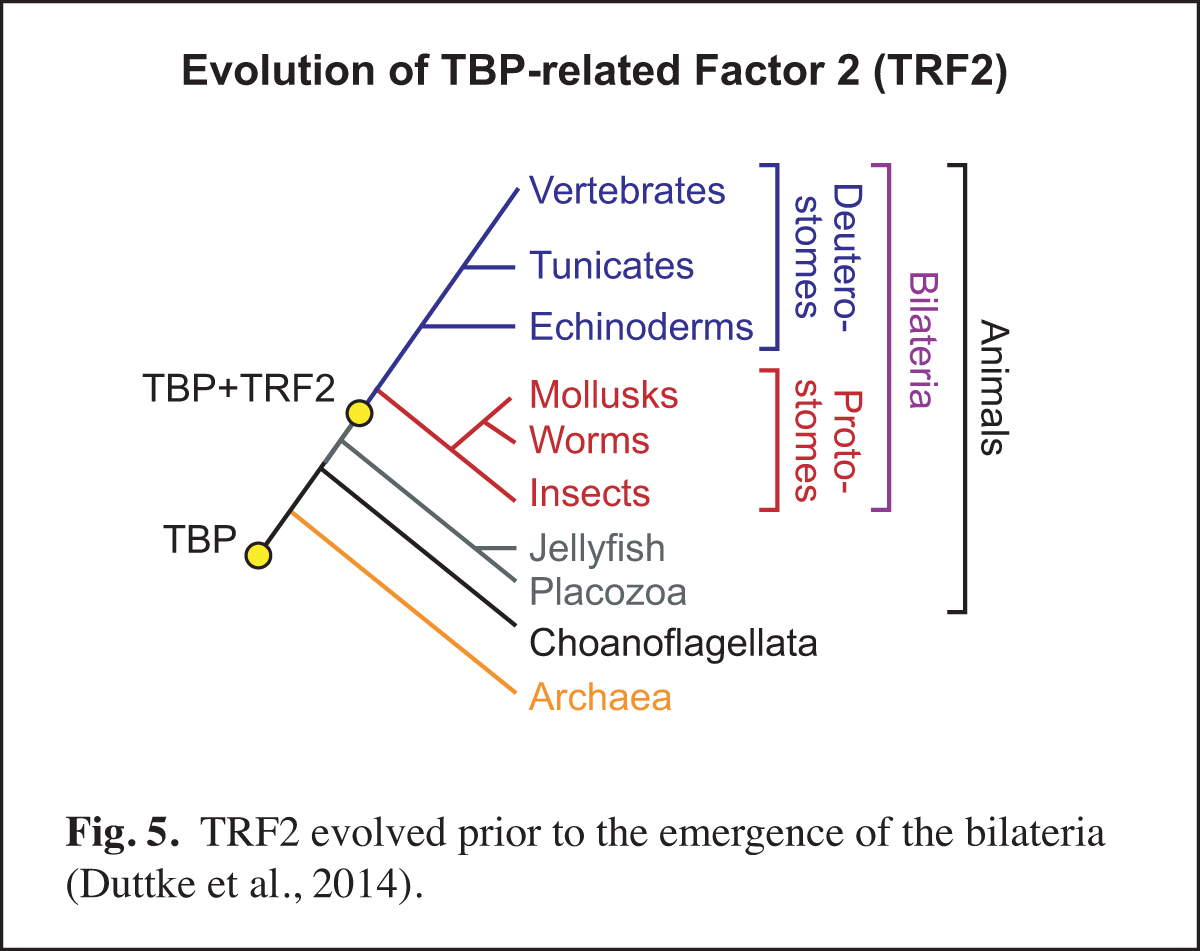
At a higher level than the DNA-based signals and their associated transcription factors, core promoter elements function in biological networks. For instance, we found that the DPE is associated with the Hox gene network and that the TCT motif is used by the ribosomal protein gene network (Fig. 4). Notably, in Drosophila, both the DPE and TCT elements function with TRF2 instead of TBP. These findings further led us to investigate the evolution of TRF2 (Fig. 5). This work suggested that the appearance of TRF2 may have facilitated the evolution of bilateria (bilaterally-symmetric animals). These studies exemplify our interest in understanding the function of the core promoter in a broader biological context.
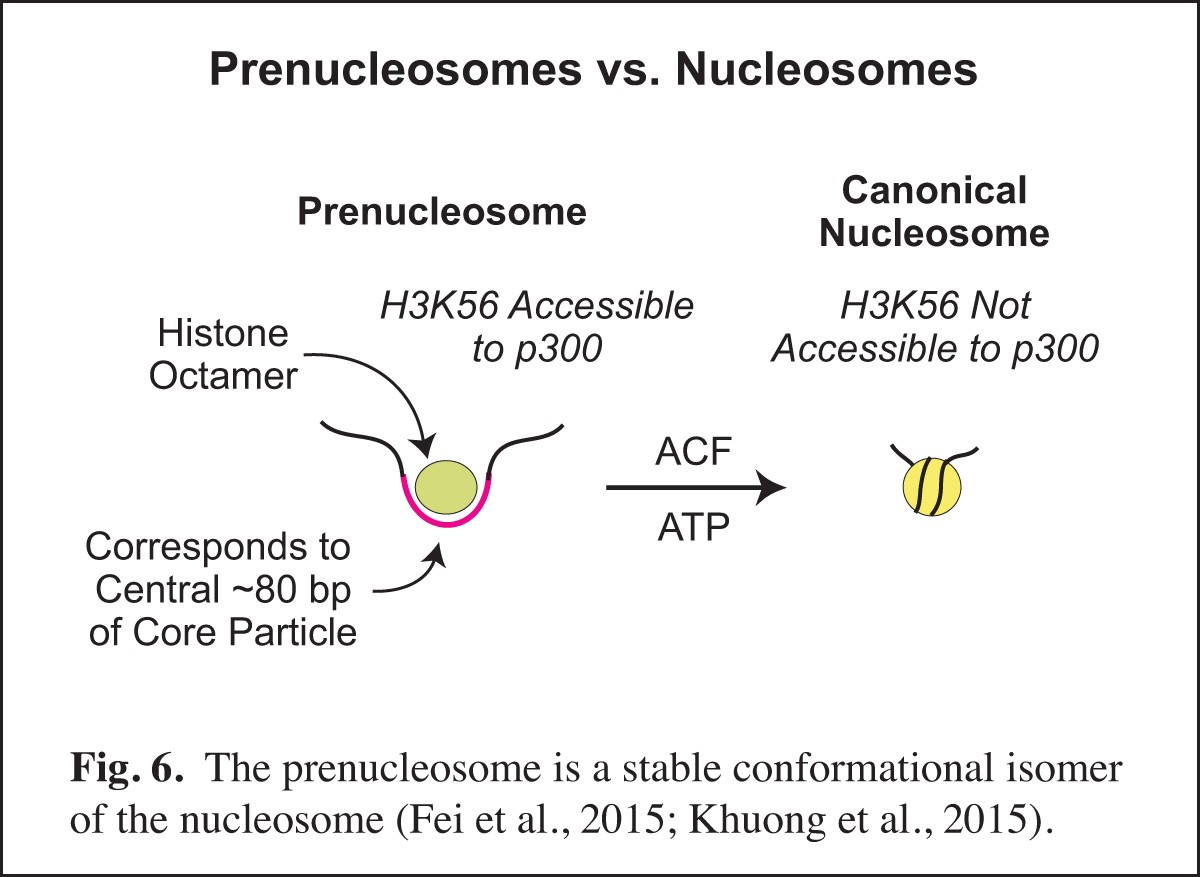
The Prenucleosome, a Stable Conformational Isomer of the Nucleosome
Chromatin consists of canonical nucleosomes as well as non-nucleosomal histone-containing particles. Noncanonical chromatin particles are typically seen in dynamic chromatin, such as at active promoters in which there is recurring disruption of nucleosomes. In the course of our studies of chromatin assembly (see Past Research below; Fig. 14), we discovered the prenucleosome, which is a stable conformational isomer of the nucleosome that associates with approximately 80 bp DNA (Fig. 6). Prenucleosomes are formed rapidly upon the deposition of histones onto DNA, and can be converted into canonical nucleosomes by an ATP-driven chromatin assembly factor such as ACF (Fig. 14). Moreover, p300 acetylates histone H3K56 in prenucleosomes but not in nucleosomes.
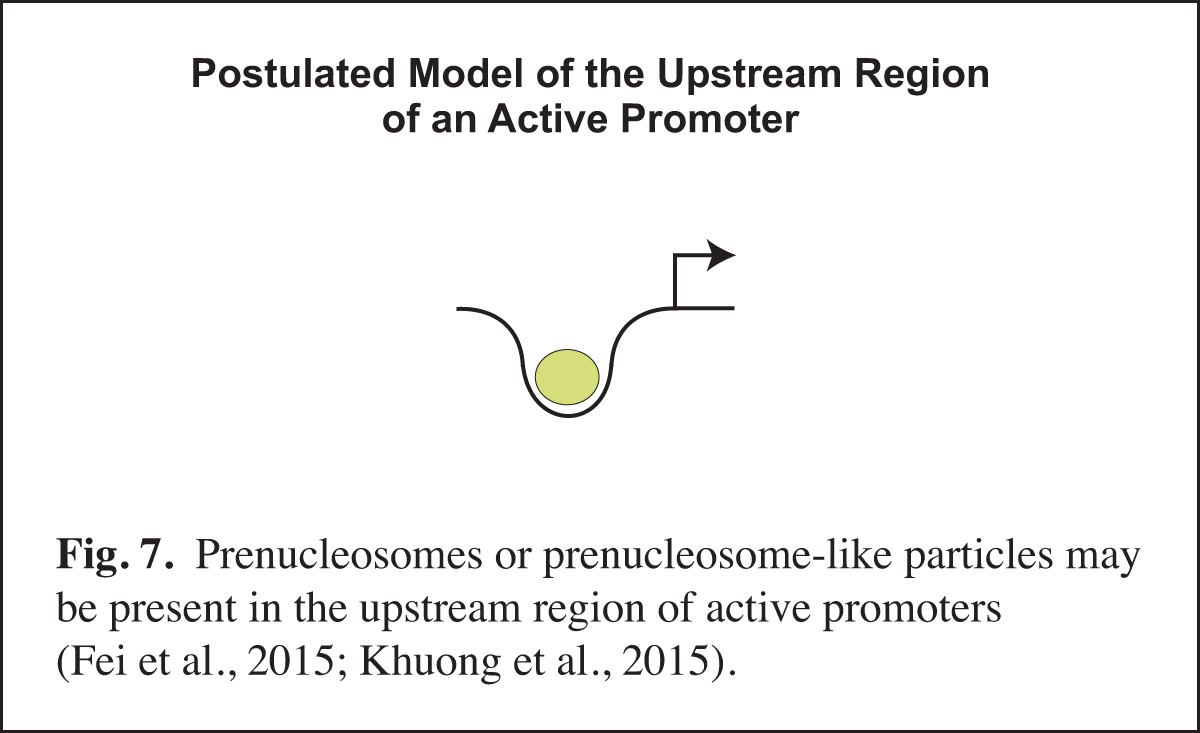
Prenucleosomes or prenucleosome-like particles appear to be present at active promoters (Fig. 7). For instance, there is a strong correlation between the DNA lengths observed with prenucleosomes assembled in vitro and the DNA lengths seen in particles at the active (but not repressed) yeast PHO5 promoter in vivo. Moreover, histone-containing chromatin particles with prenucleosome-sized DNA are observed at the upstream region of active promoters in mouse cells. The presence of prenucleosome-like particles at active promoters may reflect the recurring disruption of nucleosomes by transcription factors. It is also possible that prenucleosomes might facilitate transcription at active promoters. There is much that remains to be learned about noncanonical chromatin particles such as prenucleosomes.
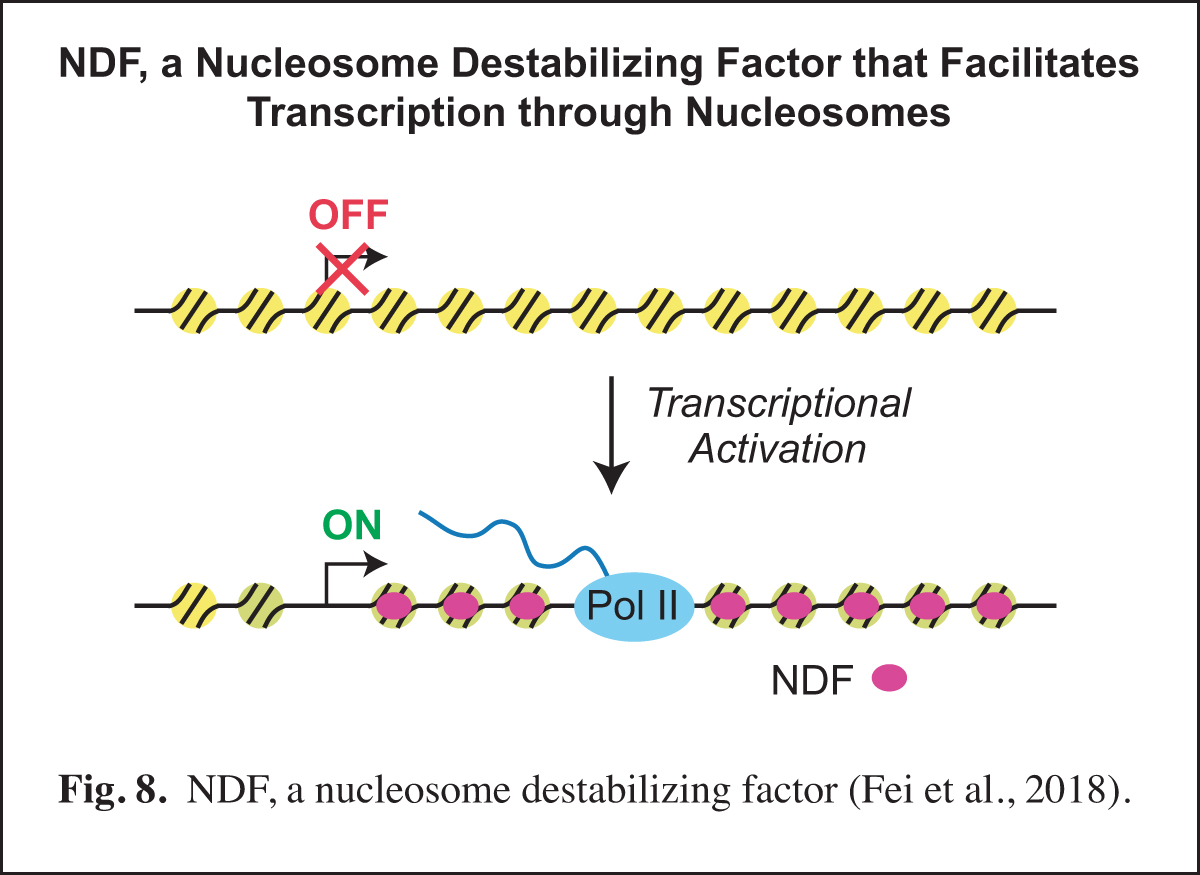
NDF, a Nucleosome Destabilizing Factor that Facilitates Transcription through Nucleosomes
Our studies of prenucleosomes inspired us to investigate factors that unwrap and/or destabilize nucleosomes. In these studies, we identified and purified NDF (also known as CG4747 in Drosophila and GLYR1, NPAC, and NP60 in humans), a nucleosome-destabilizing factor that facilitates RNA polymerase II transcription. NDF has a PWWP motif, interacts with nucleosomes near the dyad, destabilizes nucleosomes in an ATP-independent manner, and facilitates transcription by RNA polymerase II through nucleosomes in a purified and defined transcription system. Upon transcriptional induction, NDF is recruited to the transcribed regions of thousands of genes and co-localizes with a subset of H3K36me3-enriched regions. The recruitment of NDF to gene bodies is accompanied by an increase in the transcript levels of many of the NDF-enriched genes. In addition, the loss of NDF results in a decrease in the transcript levels of many genes, particularly those in which NDF is normally enriched. In humans, NDF is present at high levels in cell nuclei in all tested tissue types (44/44). These findings indicate that NDF is a nucleosome-destabilizing factor that is recruited to gene bodies during transcriptional activation and facilitates RNA polymerase II transcription (Fig. 8).
The properties of NDF indicate that it has a key role in the transcription of chromatin by RNA polymerase II in animals. The further analysis of NDF should reveal additional factors and mechanisms that orchestrate the procession of RNA polymerase II through active genes.
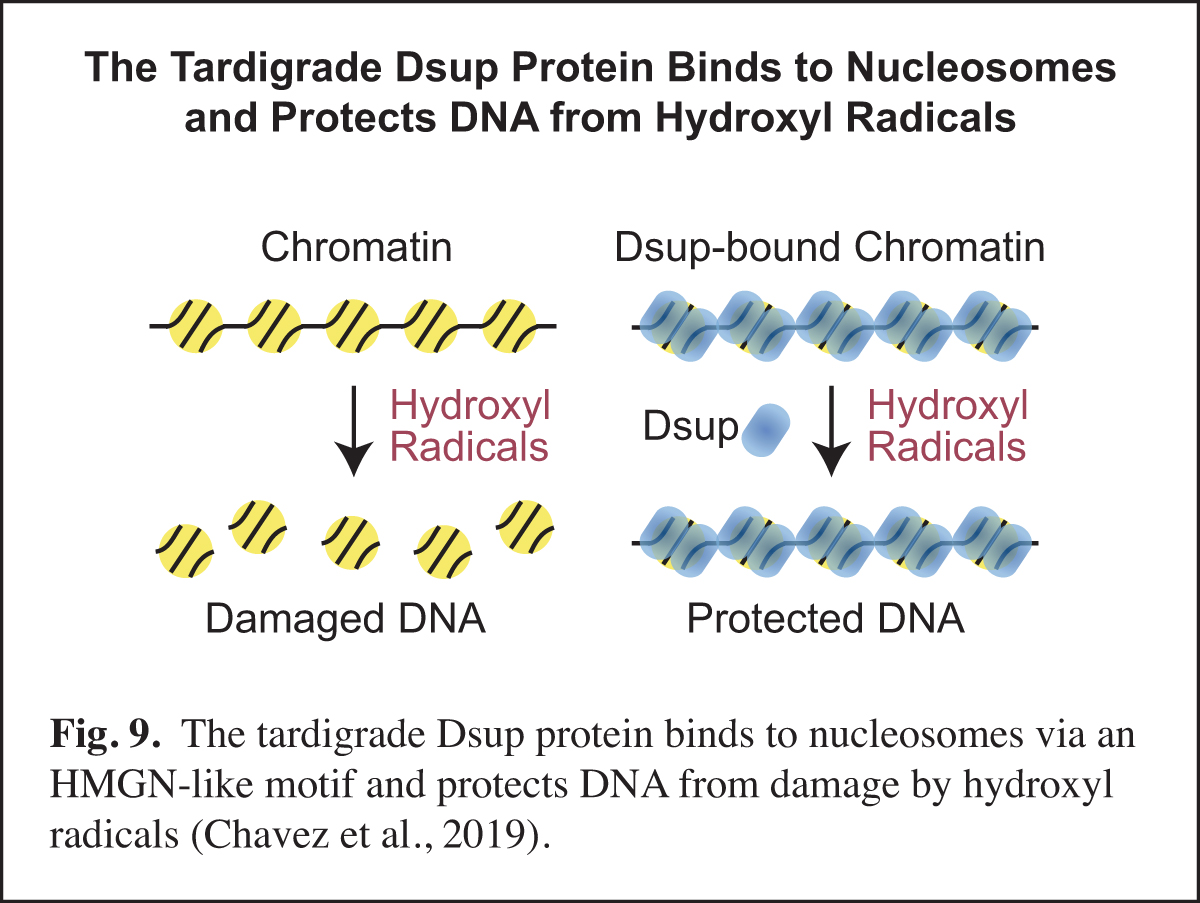
The Tardigrade Dsup Protein Binds to Nucleosomes and Protects DNA from Hydroxyl Radicals
Tardigrades, also known as water bears, are animals that can survive extreme conditions. The tardigrade Ramazzottius varieornatus contains a unique nuclear protein termed Dsup, for damage suppressor, which can increase the resistance of human cells to DNA damage under conditions (such as ionizing radiation or hydrogen peroxide treatment) that generate hydroxyl radicals. In our work, we found that R. varieornatus Dsup is a nucleosome-binding protein that protects chromatin from hydroxyl radicals. Moreover, a Dsup ortholog from the tardigrade Hypsibius exemplaris similarly binds to nucleosomes and protects DNA from hydroxyl radicals. Strikingly, a conserved region in Dsup proteins exhibits sequence similarity to the nucleosome-binding domain of vertebrate HMGN proteins and is functionally important for nucleosome binding and hydroxyl radical protection. These findings suggest that Dsup promotes the survival of tardigrades under diverse conditions by a direct mechanism that involves binding to nucleosomes and protecting chromosomal DNA from hydroxyl radicals (Fig. 9).
In this work, we thought that we might learn something new and important by studying an intriguing protein from an extreme organism. One particularly unexpected finding was the HMGN-like motif in Dsup. HMGN proteins had been previously thought to be present only in vertebrates. The discovery of the HMGN-like motif in Dsup should lead to new insights into this motif and its potential role in diverse organisms.
Scientific American PodcasteLife Podcast
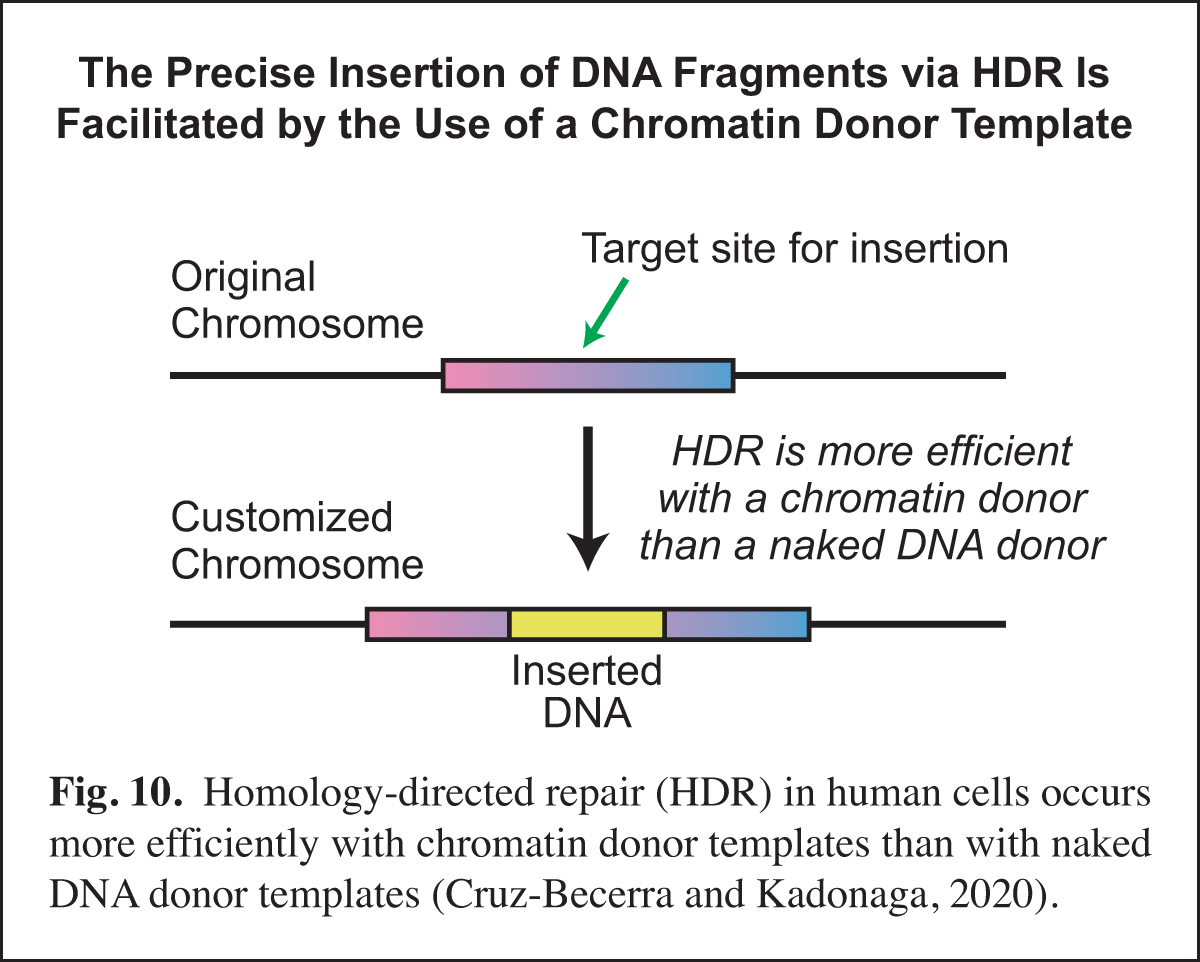
Enhancement of Homology-directed Repair (HDR) with Chromatin Donor Templates in Human Cells
In the course of our work, we will sometimes see an opportunity to contribute to research in other labs via the development of a new method. Examples of such methods include optimized core promoters (Juven-Gershon et al., 2006; Pfeiffer et al., 2008), MPE-seq (Ishii et al., 2015), and new chromatin assembly methods (Khuong et al., 2017). Recently, based on our earlier work on homologous strand pairing (Alexiadis and Kadonaga, 2002), we investigated homology-directed repair (HDR) in human cells.
A key challenge in precise genome editing is the low efficiency of HDR. We therefore developed a strategy for increasing the efficiency of HDR in cells by using a chromatin donor template instead of a naked DNA donor template. The use of chromatin, which is the natural form of DNA in the nucleus, increases the frequency of HDR-edited clones as well as homozygous editing. In addition, transfection of chromatin results in negligible cytotoxicity. These findings suggest that a chromatin donor template should be useful for a wide range of HDR applications such as the precise insertion or replacement of DNA fragments that contain the coding regions of genes (Cruz-Becerra and Kadonaga, 2020) (Fig. 10).
Past Research
Development of Sequence-specific DNA Affinity Chromatography & the Purification and Cloning of Transcription Factor Sp1
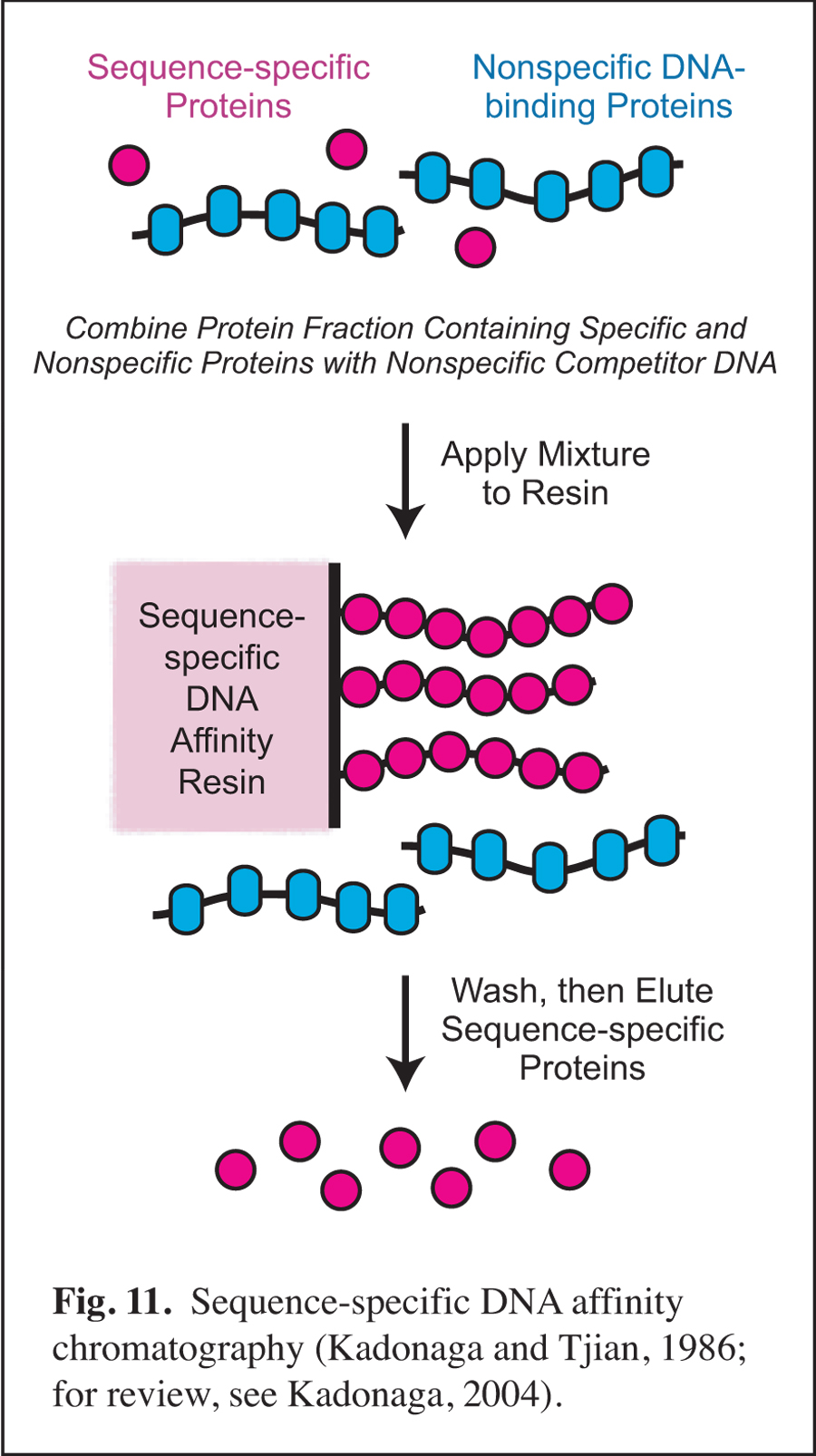
Prior to my arrival at UCSD, I was involved in early studies of eukaryotic sequence-specific DNA-binding transcriptional factors as a postdoctoral researcher in the laboratory of Robert Tjian at UC Berkeley (for review, see: Kadonaga, 2004). In this work, I developed sequence-specific DNA affinity chromatography for the purification of transcription factor Sp1 (Fig. 11). The purification of Sp1 enabled me to isolate its corresponding cDNA and to characterize the binding of Sp1 to DNA via its three zinc finger motifs (Kadonaga et al., 1987). Sequence-specific DNA affinity chromatography has since been used by many investigators to purify numerous factors that function in a wide range of biological processes.
Early Studies of the Role of Chromatin in the Regulation of Transcription
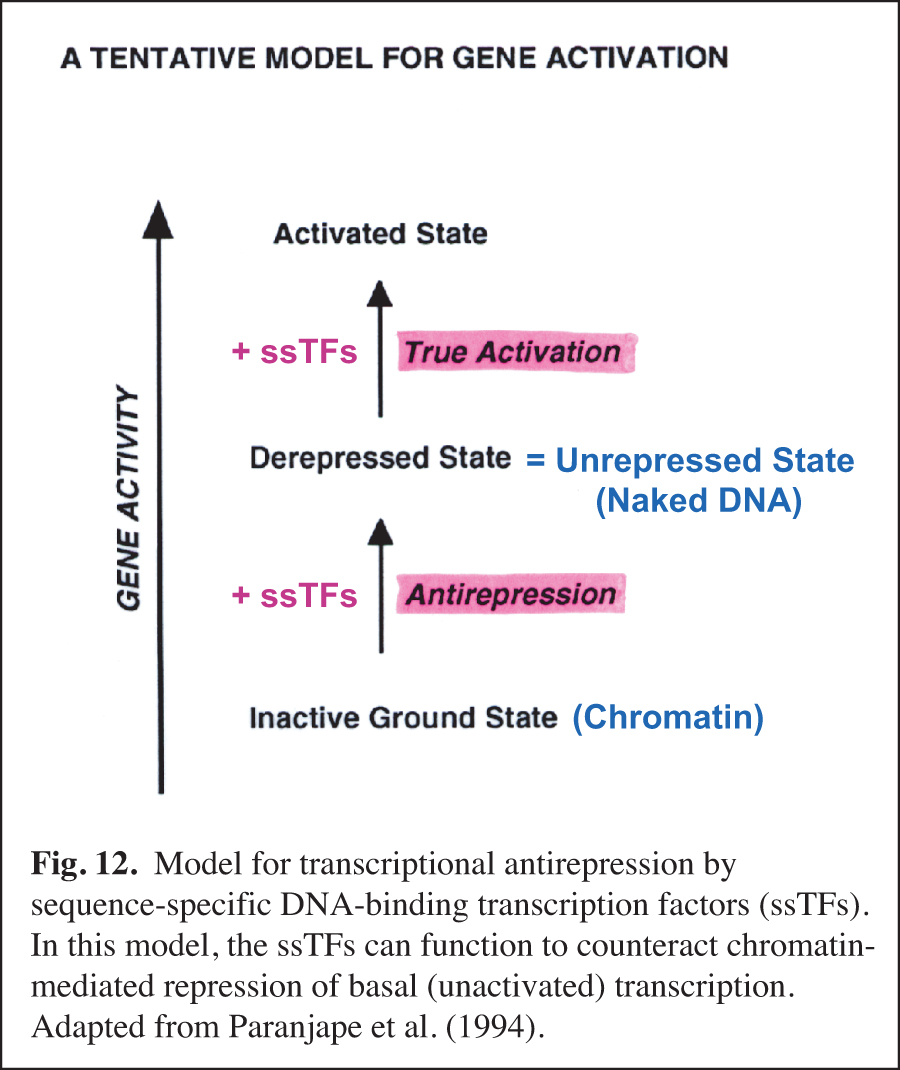
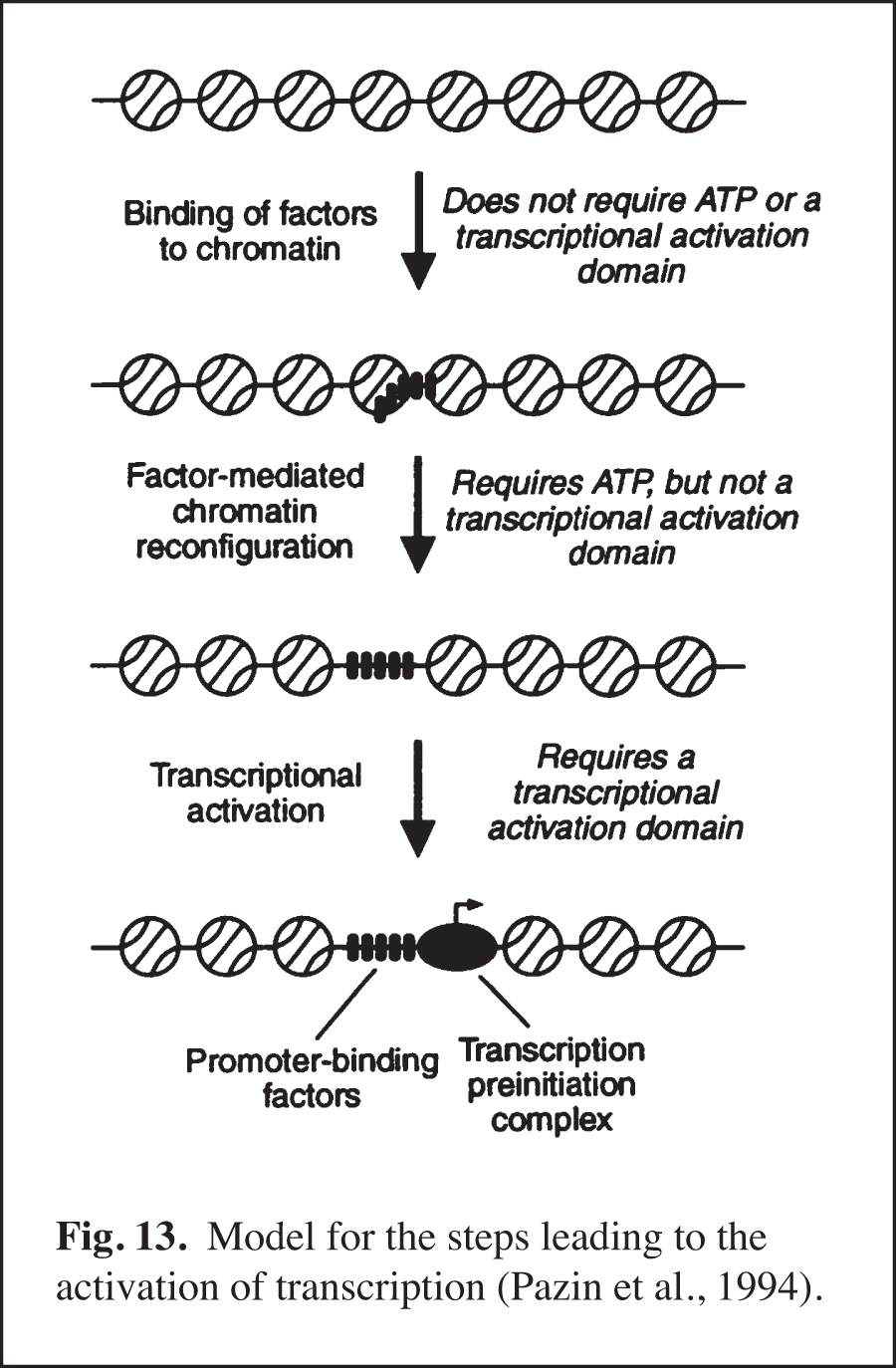
We carried out early experiments on the role of chromatin structure in the regulation of transcription by RNA polymerase II (reviewed in Kadonaga, 2019). These studies were initiated in the late 1980s – this was a time when many people in the transcription field felt that chromatin was unimportant. We were primarily motivated to perform this work because chromatin is the natural state of DNA in the eukaryotic nucleus. We found that sequence-specific DNA-binding transcription factors act primarily to counteract chromatin-mediated repression – we termed this phenomenon "antirepression" (Laybourn and Kadonaga, 1991, 1992; Paranjape et al., 1994) (Fig. 12). This emerging paradigm was in contrast to the prevailing belief that sequence-specific factors increase the rate of the transcription reaction in a manner that is independent of chromatin structure. These studies of chromatin and transcription led to the model for transcriptional activation that is shown in Fig. 13 (Pazin et al., 1994). In this project, we were also able to achieve ligand-regulated transcription in vitro by the estrogen receptor by using chromatin templates (Kraus and Kadonaga, 1998). Today, it is generally accepted that sequence-specific activators counteract chromatin-mediated repression of basal (unactivated) transcription. Chromatin is now extensively studied as an important and integral component of transcriptional regulation.
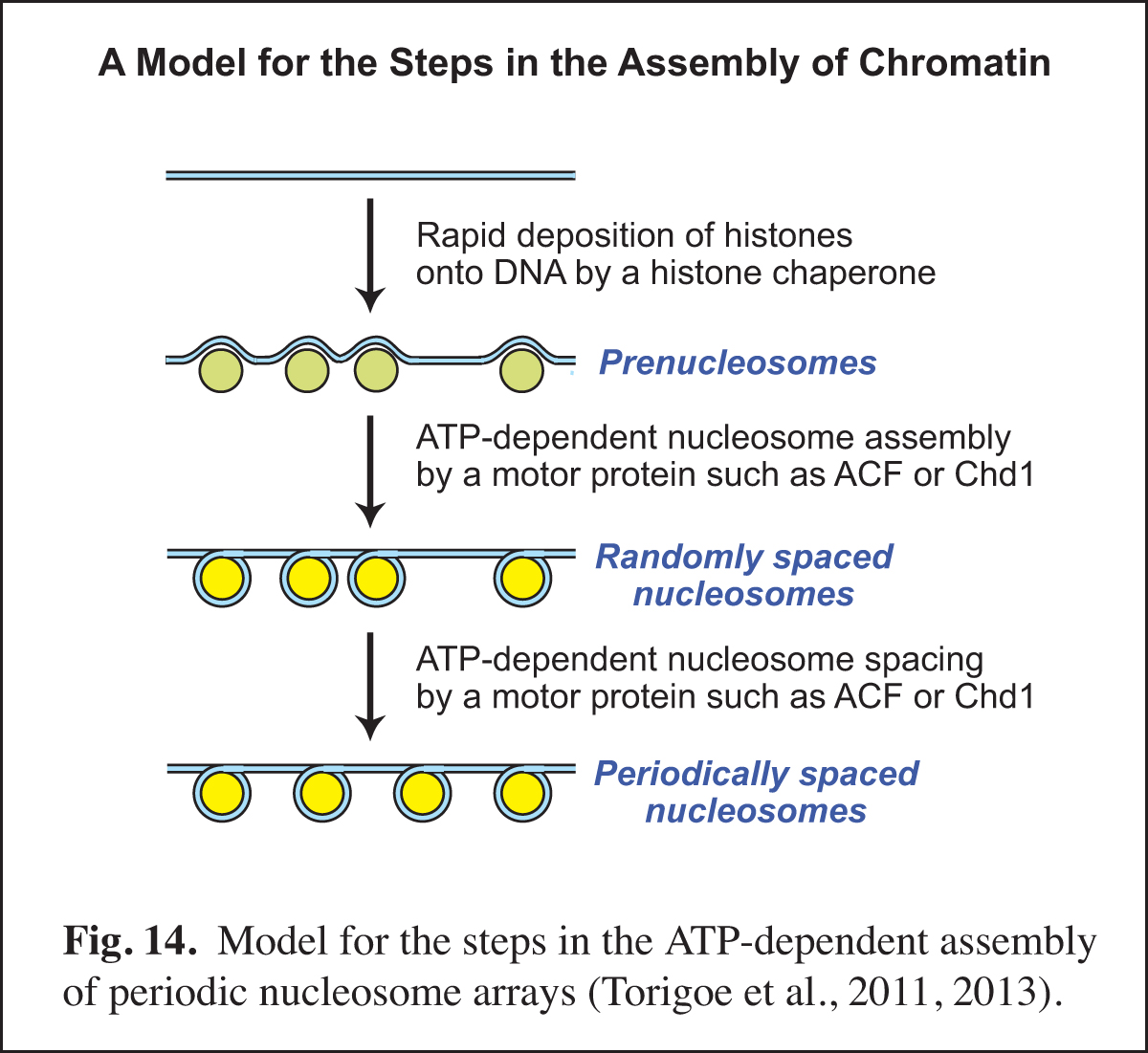
Identification of the Factors that Mediate the ATP-dependent Assembly of Chromatin
In the eukaryotic nucleus, chromatin assembly is essential for DNA (i.e., chromatin) transcription, replication, and repair as well as for the generation of specialized chromatin regions, such as those containing histone variants. Our studies of chromatin assembly began with the development of a crude extract from Drosophila embryos, termed the S-190, that is competent for ATP-dependent chromatin assembly. Over the next nine years, we carried out the biochemical fractionation and purification of the essential components in the S-190 extract. This work included the discovery, purification, and cloning of ACF (ATP-utilizing chromatin assembly and remodeling factor; Ito et al., 1997, 1999) and the discovery that Asf1 is a chromatin assembly factor that serves as a chaperone for newly synthesized histones H3 and H4 (Tyler et al., 1999). The analysis of the mechanism of the ATP-dependent assembly of periodic nucleosome arrays led to the model shown in Fig. 14.
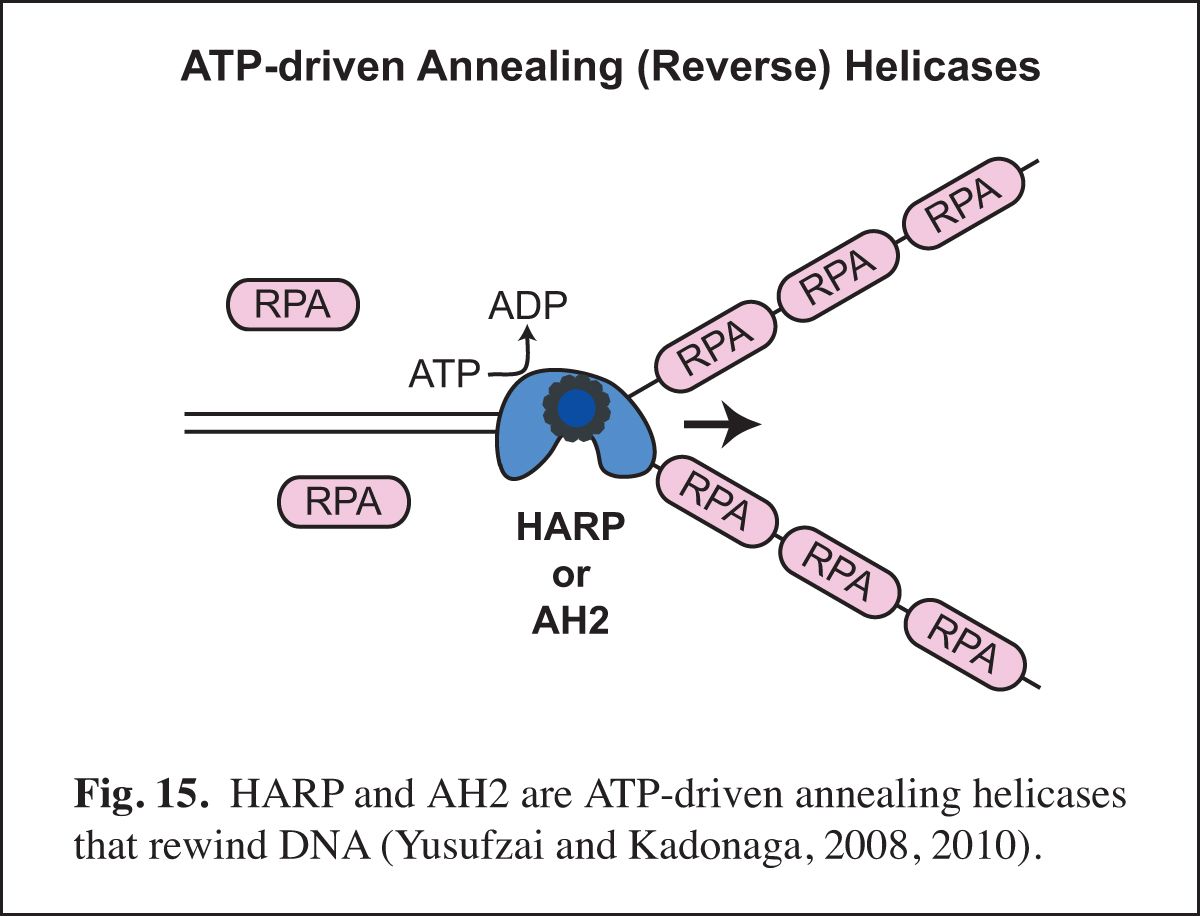
Identification of Annealing Helicases
Our studies of chromatin dynamics took an unexpected turn in the course our studies of the HARP protein (also known as SMARCAL1). We were interested in HARP because it is in the same family of ATPases (the SNF2-like family) as the chromatin assembly proteins ISWI (the ATPase subunit of ACF) and Chd1. In addition, mutations in HARP were found to be responsible for a rare human disorder known as Schimke immuno-osseous dysplasia (SIOD). We embarked on the analysis of HARP with the notion that it might alter chromatin structure; however, HARP did not exhibit chromatin remodeling activity. Instead, we discovered that HARP possesses a novel annealing (reverse) helicase activity. HARP does not unwind DNA like DNA helicases. Thus, HARP is a motorized molecular zipper that anneals DNA, such as complementary single DNA strands that are bound by RPA, the major single-stranded DNA-binding protein in the nucleus (Fig. 15). In addition, we found a second annealing helicase termed annealing helicase 2 (AH2, also known as ZRanB3). Unlike HARP, which forms a stable complex with RPA, AH2 does not bind to RPA. It is particularly notable that HARP and AH2 function only to anneal DNA and do not possess DNA-unwinding activity like conventional helicases. In this manner, they act in opposition to conventional helicases and other enzymes that unwind DNA.
Selected Publications
- Fei, J.*, Xu, J., Li, Z., Xu, K., Wang, D., Kassavetis, G. A., and Kadonaga, J. T.* (2022). NDF is a transcription factor that stimulates elongation by RNA polymerase II. Genes Dev. 36, 294-299. *Co-corresponding authors. PMCID: PMC8973848
- Vo ngoc, L., Huang, C. Y., Cassidy, C. J., Medrano, C., and Kadonaga, J. T. (2020). Identification of the human DPR core promoter element using machine learning. Nature 585, 459-363. PMCID: PMC7501168
- Cruz-Becerra, G., and Kadonaga, J. T. (2020). Enhancement of homology-directed repair with chromatin donor templates in cells. eLife 9, e55780. PMCID: PMC7244325
- Chavez, C.†, Cruz-Becerra, G. †, Fei, J. †, Kassavetis, G. A. †, and Kadonaga, J. T. (2019). The tardigrade damage suppressor protein binds to nucleosomes and protects DNA from hydroxyl radicals. eLife 8, e47682. †Co-first authors. PMCID: PMC6773438
- Kadonaga, J. T. (2019). The transformation of the DNA template in RNA polymerase II transcription: a historical perspective. Nat. Struct. Mol. Biol. 26, 766-770. PMCID: PMC7025952
- Vo ngoc, L., Kassavetis, G. A., and Kadonaga, J. T. (2019). The RNA polymerase II core promoter in Drosophila. Genetics 212, 13-24. *Contribution to the Drosophila FlyBook. PMCID: PMC6499525
- Fei, J., Ishii, H., Hoeksema, M. A., Meitinger, F., Kassavetis, G. A., Glass, C. K., Ren, B., and Kadonaga, J. T. (2018). NDF, a nucleosome destabilizing factor that facilitates transcription through nucleosomes. Genes Dev. 32, 682-694. PMCID: PMC6004073
- Khuong, M.T., Fei, J., Cruz-Becerra, G., and Kadonaga, J.T. (2017). A simple and versatile system for the ATP-dependent assembly of chromatin. J. Biol. Chem. 292, 19478-19490. PMCID: PMC5702684
- Vo ngoc, L., Wang, Y.-L., Kassavetis, G.A., and Kadonaga, J.T. (2017). The punctilious RNA polymerase II core promoter. Genes Dev. 31, 1289-1301. PMCID: PMC5580651
- Vo ngoc, L., Cassidy, C.J., Huang, C.Y., Duttke, S.H.C., and Kadonaga, J.T. (2017). The human initiator is a distinct and abundant element that is precisely positioned in focused core promoters. Genes Dev. 31, 6-11. PMCID: PMC5287114
- Fei, J., Torigoe, S.E., Brown, C.R., Khuong, M.T., Kassavetis, G.A., Boeger, H., and Kadonaga, J.T. (2015). The prenucleosome, a stable conformational isomer of the nucleosome. Genes Dev. 29, 2563-2575. PMCID: PMC4699385
- Khuong, M.T., Fei, J., Ishii, H., and Kadonaga, J.T. (2015). Prenucleosomes and active chromatin. Cold Spring Harbor Symp. Quant. Biol. 80, in press. doi: 10.1101/sqb.2015.80.027300
- Duttke, S.H.C., Lacadie, S.A., Ibrahim, M.M., Glass, C.K., Corcoran, D.L., Benner, C., Heinz, S., Kadonaga, J.T.‡, and Ohler, U.‡ (2015). Human promoters are intrinsically directional. Mol. Cell 57, 674-684. ‡Co-corresponding authors. PMCID: PMC4336624
- Ishii, H., Kadonaga, J. T. ‡, and Ren, B. ‡ (2015). MPE-seq, a new method for the genome-wide analysis of chromatin structure. Proc. Natl. Acad. Sci. USA 112, E3457-E3465. ‡Co-corresponding authors. PMCID: PMC4500276
- Duttke, S. H. C., Doolittle, R. F., Wang, Y.-L., and Kadonaga, J. T. (2014). TRF2 and the evolution of the bilateria. Genes Dev. 28, 2071-2076. PMCID: PMC4180970
- Wang, Y.-L., Duttke, S. H. C., Chen, K., Johnston, J., Kassavetis, G. A., Zeitlinger, J., and Kadonaga, J. T. (2014). TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 28, 1550-1555. PMCID: PMC4102762
- Torigoe, S. E., Patel, A., Khuong, M. T., Bowman, G. D., and Kadonaga, J. T. (2013). ATP-dependent chromatin assembly is functionally distinct from chromatin remodeling. eLife 2, e00863. PMCID: PMC3748710
- Kadonaga, J. T. (2012). Perspectives on the RNA polymerase II core promoter. WIREs Dev. Biol. 1, 40-51. PMCID: PMC3695423
- Torigoe, S. E, Urwin, D. L., Ishii, H., Smith, D. E., and Kadonaga, J. T. (2011). Identification of a rapidly formed non-nucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol. Cell 43, 638-648. PMCID: PMC3160715
- Yusufzai, T., and Kadonaga, J. T. (2010). Annealing helicase 2 (AH2), a DNA-rewinding motor with an HNH motif. Proc. Natl. Acad. Sci. USA 107, 20970-20973. PMCID: PMC3000258
- Parry, T. J., Theisen, J. W. M., Hsu, J.-Y., Wang, Y.-L., Corcoran, D. L., Eustice, M., Ohler, U., and Kadonaga, J. T. (2010). The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 24, 2013-2018. PMCID: PMC2939363
- Juven-Gershon, T., and Kadonaga, J. T. (2010). Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225-229. PMCID: PMC2830304
- Yusufzai, T., Kong, X., Yokomori, K., and Kadonaga, J. T. (2009). The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 23, 2400-2404. PMCID: PMC2764493
- Rattner, B. P., Yusufzai, T., and Kadonaga, J. T. (2009). HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Mol. Cell 34, 620-626. PMCID: PMC2709789
- Yusufzai, T., and Kadonaga, J. T. (2008). HARP is an ATP-driven annealing helicase. Science 322, 748-750. PMCID: PMC2587503
- Juven-Gershon, T., Hsu, J.-Y., and Kadonaga, J. T. (2008). Caudal, a key developmental regulator, is a DPE-specific transcription factor. Genes Dev. 22, 2823-2830. PMCID: PMC2569877
- Hsu, J.-Y., Juven-Gershon, T., Marr, M. T., 2nd, Wright, K. J., Tjian, R., and Kadonaga, J. T. (2008). TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 22, 2353-2358. PMCID: PMC2532932
- Santoso, B., and Kadonaga, J. T. (2006). Reconstitution of chromatin transcription with purified components reveals a chromatin-specific repressive activity of p300. Nature Struct. Mol. Biol. 13, 131-139.
- Lusser, A., Urwin, D. L., and Kadonaga, J. T. (2005). Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nature Struct. Mol. Biol. 12, 160-166.
- Lim, C. Y., Santoso, B., Boulay, T., Dong, E., Ohler, U., and Kadonaga, J. T. (2004). The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18, 1606-1617.
- Kadonaga, J. T. (2004). Regulation of RNA polymerase II transcription by sequence-specific DNA-binding factors. Cell 116, 247-257.
- Fyodorov, D. V., Blower, M. D., Karpen, G. H., and Kadonaga, J. T. (2004). Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 18, 170-183.
- Alexiadis, V., and Kadonaga, J. T. (2002). Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16, 2767-2771.
- Butler, J. E. F., and Kadonaga, J. T. (2002). The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16, 2583-2592.
- Fyodorov, D. V., and Kadonaga, J. T. (2002). Dynamics of ATP-dependent chromatin assembly by ACF. Nature 418, 897-900.
- Butler, J. E. F., and Kadonaga, J. T. (2001). Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515-2519.
- Willy, P. J., Kobayashi, R., and Kadonaga, J. T. (2000). A basal transcription factor that activates or represses transcription. Science 290, 982-984.
- Tyler, J. K., Adams, C. R., Chen, S.-R., Kobayashi, R., Kamakaka, R. T., and Kadonaga, J. T. (1999). The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555-560.
- Ito, T., Levenstein, M. E., Fyodorov, D. V., Kutach, A. K., Kobayashi, R., and Kadonaga, J. T. (1999). ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13, 1529-1539.
- Blackwood, E. M., and Kadonaga, J. T. (1998). Going the distance: a current view of enhancer action. Science 281, 60-63.
- Kraus, W. L., and Kadonaga, J. T. (1998). p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12, 331-342.
- Burke, T. W., and Kadonaga, J. T. (1997). The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11, 3020-3031.
- Ito, T., Bulger, M., Pazin, M. J., Kobayashi, R., and Kadonaga, J. T. (1997). ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145-155.
- Pazin, M. J., and Kadonaga, J. T. (1997). What's up and down with histone deacetylation and transcription? Cell 89, 325-328.
- Pazin, M. J., Bhargava, P., Geiduschek, E. P., and Kadonaga, J. T. (1997). Nucleosome mobility and the maintenance of nucleosome positioning. Science 276, 809-812.
- Pazin, M. J., and Kadonaga, J. T. (1997). SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88, 737-740.
- Burke, T. W., and Kadonaga, J. T. (1996). Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10, 711-724.
- Pazin, M. J., Kamakaka, R. T., and Kadonaga, J. T. (1994). ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science 266, 2007-2011.
- Paranjape, S. M., Kamakaka, R. T., and Kadonaga, J. T. (1994). Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu. Rev. Biochem. 63, 265-297.
- Kamakaka, R. T., Bulger, M., and Kadonaga, J. T. (1993). Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 7, 1779-1795.
- Tyree, C. M., George, C. P., Lira-DeVito, L. M., Wampler, S. L., Dahmus, M. E., Zawel, L., and Kadonaga, J. T. (1993). Identification of minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 7, 1254-1265.
- Laybourn, P. J., and Kadonaga, J. T. (1992). Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science 257, 1682-1685.
- Laybourn, P. J., and Kadonaga, J. T. (1991). Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254, 238-245.
- Croston, G. E., Kerrigan, L. A., Lira, L., Marshak, D. R., and Kadonaga, J. T. (1991). Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science 251, 643-649.
- Kamakaka, R. T., Tyree, C. M., and Kadonaga, J. T. (1991). Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proc. Natl. Acad. Sci. USA 88, 1024-1028.
- Kadonaga, J. T., Courey, A. J., Ladika, J., and Tjian, R. (1988). Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science 242, 1566-1570.
- Kadonaga, J. T., Carner, K. R., Masiarz, F. R., and Tjian, R. (1987). Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51, 1079-1090.
- Kadonaga, J. T., and Tjian, R. (1986). Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. USA 83, 5889-5893.
Biography
James Kadonaga was an undergraduate in chemistry at MIT, where he performed undergraduate research with Rick L. Danheiser and received both the Alpha Chi Sigma Prize and the American Institute of Chemists Certificate in 1980. He carried out his graduate studies (1980-84) with Jeremy R. Knowles in the Department of Chemistry at Harvard University, where he was a DuPont Fellow. Jim was a postdoctoral associate (1984-88) with Robert Tjian at UC Berkeley as a Fellow of the Miller Institute for Basic Research in Science, American Cancer Society California Division, and Lucille P. Markey Charitable Trust. He joined the faculty of UCSD in 1988 and was one of 15 scientists to be named as a Presidential Faculty Fellow by President George H.W. Bush in 1992. In 1994, he was elected to Fellow of the American Association for the Advancement of Science, and in 1995, he was elected to Fellow of the American Academy of Microbiology. He served as the Chair of the Department (formerly Section) of Molecular Biology from 2003 to 2007. In 2012, he received the UCSD Chancellor's Associates Award for Excellence in Research in Science and Engineering. In 2017, he was elected to the American Academy of Arts & Sciences. In 2020, he was elected to Revelle College Faculty Fellow at UCSD. He was elected to the National Academy of Sciences in 2022. Jim is presently the Amylin Endowed Chair and a Distinguished Professor in the Department of Molecular Biology.
Profile in Journal of Cell Biology.
Personal retrospective article in Nature Structural & Molecular Biology.
Q&A interview in the 25th Anniversary Issue of Molecular Cell.
Letters to the Editor in the Special Terri Grodzicker Issue of Genes & Development

