Justin Meyer
Research
The Meyer Lab at UC San Diego addresses some of the most challenging questions in evolutionary theory, such as speciation, novelty, evolvability, and complexity. The team designs experiments to observe these processes in real time. Using trackable microbial platforms and systems and synthetic biology, they generate new datasets and test evolutionary and ecological theories that were previously untestable. Their work often focuses on Bacteriophage λ, which evolves to exploit alternative receptors. This expansion of host range provides key insights into host-pathogen interactions.
The lab is also advancing phage therapy through innovative approaches. They develop "evolutionary training" protocols to pre-evolve bacteriophages, helping them counter bacterial resistance. The team collaborates on synthetic phage platforms, including gene drive technologies, to remove antibiotic resistance genes from microbiomes. These efforts resensitize bacteria and restore antibiotic effectiveness. By linking theory with practical applications, the lab drives innovation in microbial evolution and phage therapeutics.
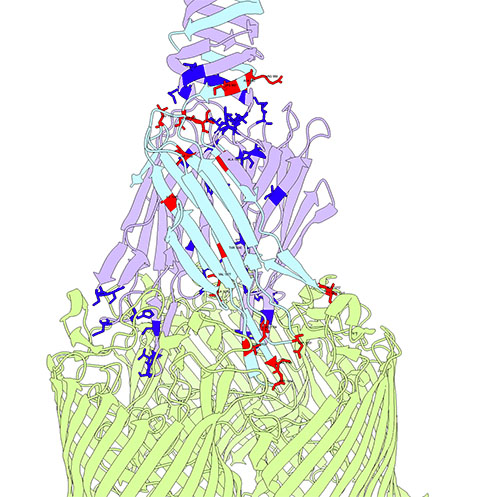
Bacteriophage λ's receptor-binding protein interacts with its receptor, with key evolutionary hotspots highlighted.
Select Publications
- Borin, J.M., Lee, J.L., Lucia-Sanz, A., Gerbino, K.R., Weitz, J.W., Meyer, J.R. (2023) Rapid bacteria-phage coevolution drives the emergence of multiscale networks Science 382, 6671.
- Borin, J.M., Avrani, S., Barrick, J.E., Petrie, K.L., Meyer, J.R. (2021) Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance Proceedings of the National Academy of Sciences 23, 118.
- Petrie, K.P., Palmer, N.D., Johnson, D.T., Medina, S.J., Yan, S.J., Li, V., Burmeister, A.R., Meyer, J.R. (2018) Destabilizing mutations encode nongenetic variation that drives evolutionary innovation. Science 359: 1542-1545.
- Meyer, J.R., Dobias, D.T., Medina, S.J., Servilio, L., Gupta, A. and Lenski, R.E. (2016) Ecological speciation of bacteriophage lambda in allopatry and sympatry. Science, 354,1301-1304.
- Meyer, J.R. , Dobias, D.T., Weitz, j. S. Barrick, J.E. , Quick, R.T., Lenski R.E. (2012) Repeatability and contingency in the evolution of a key innovation in Lambda Phage. Science 335, 428-432.
- Flores, C., Meyer, J.R., Valverde, S., Farr, L., Weitz, J.S. (2011) Statistical structure of host-phage interactions Proceedings of the National Academy of Sciences 108, 28.
Biography
Justin Meyer received his Ph.D. from Michigan State University and was a Systems Biology Departmental Fellow at Harvard Medical School, where he was awarded the James S. McDonnell Foundation Fellowship for Studying Complex Systems. He joined the faculty of Ecology Behavior and Evolution and the Quantitative Biology Initiative in 2014.

