Kit Pogliano
Research
Our research is organized around three key themes:
- Cellular organization and dynamics: What are the design principles of bacterial cells?
- Microbial interactions: How are the outcomes of interspecies interactions determined?
- Chemical Cell Biology: How do secreted metabolites and antibiotics affect bacterial cells?
Our toolkit
We take a multidisciplinary approach to our studies, using the tools of genetics, cell biology, proteomics, biochemistry, and we have recently added analytical chemistry to our toolkit. Our lab is fully equipped for molecular biology, genetics and for work with BSL2 pathogens. Our microscopy facility includes one dissecting stereomicroscope equipped for fluorescence and timelapse imaging as well as deconvolution microscopes equipped with growth chambers and microfluidic devices for time-lapse microscopy and lasers for photobleaching and total internal reflection microscopy. We have an HPLC apparatus for the analysis and purification of natural products, and more extensive equipment is available in the labs of our campus collaborators. We have ready access to excellent campus facilities that provide cutting edge equipment and analytical tools for protein and small molecule mass spectrometry, super resolution microscopy and sequencing, with excellent technical assistance available as needed. We also apply our imaging expertise to a variety of collaborative projects and enjoy the diversity these projects and visitors bring to our lab.
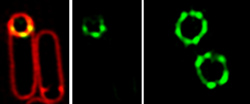
The SpollQ landmark protein (green) localizes into puncta surrounding the developing spore, here visualized using structured illumination microscopy.
Cellular organization and dynamics: What are the design principles of bacterial cells?
The phagocytosis-like process of engulfment is a hallmark of endospore formation in Bacillus subtilis and its relatives. Engulfment provides a unique example of phagocytosis in a bacterium and it is an ideal system to study protein localization and dynamics as well as membrane movement and fission. Engulfment depends on three protein complexes, the DMP complex that degrades peptidoglycan, the Q-AH complex that provides a Velcro-like attachment between the two cells and the peptidoglycan biosynthetic complex. Our goal is to elucidate the mechanisms by which bacterial cells mediate membrane movement and membrane fission, coordinate peptidoglycan degradation and synthesis, and how they complete membrane fission after engulfment and cell division.
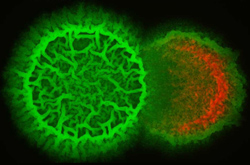
B. subtilis (green) invading and destroying a neighboring bacterial colony (red).
What role do antibiotics play in microbial communities?
The vast majority of commercial antibiotics are derived from secreted bacterial natural products. However, despite the clinical importance of these molecules and the pressing need to discover new antibiotics, it remains unclear exactly what role these molecules play in nature. The secreted metabolome of B. subtilis contains a wide array of well-characterized antibacterial and antifungal compounds, providing an ideal starting point to elucidate the specific roles each play in interspecies interactions and development. Undomesticated B. subtilis strains have the ability to invade and destroy neighboring colonies or to form mixed biofilms in a more cooperative mode of growth. Our goal is to identify the genes and secreted metabolites that mediate interspecies interactions, using the tools of analytical chemistry and imaging mass spectrometry (in collaboration with Pieter Dorrestein) as well as genetics and cell biology.
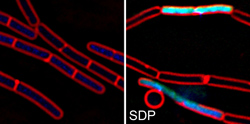
B. subtilis cells before and after treatment with the cannibalism toxin SDP.
Chemical Cell Biology: How do secreted metabolites and antibiotics affect bacterial cells?
We have developed cell biological tools to characterize the cellular targets for secreted metabolites and antibiotics, in collaboration with Joe Pogliano. This approach, which we call cytological profiling, has allowed us to rapidly identify molecules that kill bacterial cells and to determine their likely mechanism of action. It is remarkably specific, as we can discriminate between the cytological effects of molecules that dissipate either the pH or or both components of the PMF, as well as the effects of a wide array of antibiotics with well-characterized activities. These studies provide insight into the mechanisms by which essential biological processes are coordinated to maintain the appropriate size and they have several biomedical applications.
Select Publications
- Peters, C., A. Lamsa, R.B. Liu, D.T. Quach, J. Sugie, J. Pogliano, J. López -Garrido and K. Pogliano. (2018) Rapid Inhibition Profiling identifies a keystone target in the nucleotide biosynthesis pathway. Journal of Antimicrobial Chemotherapy, Aug 22. doi: 10.1021/acschembio.8b00273. PMID: 30133247
- Riley, E.P., A. Trinquier, M.L. Reilly, M. Durchon, R. Perera, K. Pogliano and J. López-Garrido. (2018) Spatiotemporally regulated proteolysis to dissect the role of vegetative proteins during Bacillus subtilis sporulation: cell-specific requirement of σ H and σ A. Molecular Microbiology 108:45-62. PMID: 29363854.
- López-Garrido, J., N. Ojkic, K. Khanna, E. Villa, R.G. Endres and K. Pogliano. (2018) Chromosome translocation inflates the Bacillus subtilis forespore and impacts cellular morphology. Cell, 172:758-770. PMID: 29425492
- Ojkic, N. López-Garrido, J., K. Pogliano and R.G. Endres. Cell wall remodeling drives engulfment during Bacillus subtilis sporulation, eLife, Nov. 17, e18657. PMID: 27852437
- Lamsa, A., J. López-Garrido, D.T. Quach, E.P. Riley, J. Pogliano and K. Pogliano. (2016) Rapid inhibition profiling in Bacillus subtilis to identify the mechanism of action of new antimicrobials, American Chemical Society Chemical Biology, 11:2222-2231. PMID: 27193499
- Nguyen, K.B., A. Sreelatha, E. Durrant, J. López-Garrido, A. Muszewska, M. Dudkiewicz, M. Grynberg, S. Yee, K. Pogliano, D. Tomchick, K. Pawłowski, J.E. Dixon and V.S. Tagliabracci. (2016) Phosphorylation of spore coat proteins by a new family of kinases. Proc. Natl. Acad. Sci., USA 113:E3482-3491. PMID: 27185916
- Nonejuie, G.L., R. Trial, G.L. Newton, A. Lamsa, V.R. Perera, J. Aguilar, W.-T. Liu, P.C. Dorrestein, J. Pogliano and K. Pogliano. (2015) Application of bacterial cytological profiling to crude natural product extracts reveals the antibacterial arsenal of Bacillus subtilis, Journal of Antibiotics. Dec 9. doi: 10.1038/ja.2015.116. PMID: 26648120.
- Shin, J.Y., J. López-Garrido, S.-H. Lee, C. Diaz, T. Fleming, C. Bustamante and K. Pogliano. (2015) Visualization and functional dissection of coaxial paired SpoIIIE channels across the sporulation septum. eLife, May 7;4. doi: 10.7554. PMID: 25950186
- Nonejuie P., M. Burkhart, K. Pogliano and J. Pogliano. (2013) Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci., USA 110:16169-16174. PMID:24046367
- Lamsa, A., W.-T. Liu, P.C. Dorrestein and K. Pogliano. (2012) The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Molecular Microbiology 84:486-500. PMID: 22469514
- Fleming, T., J.Y. Shin, S.H. Lee, E.C. Becker, K.C. Huang, C. Bustamante and K. Pogliano. (2010) Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes and Development 24:1160-1172. PMID:20516200
- Chiba, S. A. Lamsa and K. Pogliano. (2009) A ribosome nascent chain sensor of membrane protein biogenesis in Bacillus subtilis, EMBO J. 28:3461-3475 PMID: 19779460
- Gregory, J.A., E.C. Becker and K. Pogliano. (2008) Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes and Development 22:3475-3488. PMID: 19141479
- Broder, D. and K. Pogliano. (2006) Forespore engulfment mediated by a ratchet-like mechanism. Cell 126:917-928. PMID: 16959571
Biography
Kit Pogliano received her Ph.D. from the Department of Microbiology and Molecular Genetics at Harvard Medical School and was a Damon Runyon-Walter Winchell postdoctoral fellow at Harvard University. She is a recipient of the Searle Scholar and Beckman Young Investigator awards. Dr. Pogliano was named in 2009 as a Fellow in the American Academy of Microbiology.

