Vu Nguyen
Research
The cell maintains its state and type by expressing proper levels of the right genes at the right time. In the nucleus, such specificity demands constant actions by numerous factors that organize and transcribe the genome, much of which is condensed in "closed" chromatin. These factors must search the nuclear space and compete for binding to DNA in "open" chromatin. Timing is of the essence here—individual components must arrive in time to form complexes that must coordinate their functions sequentially to transcribe the target gene. In the confined and highly crowded cell nucleus, how is the genome organized and how do factors find the right genes at the right time?
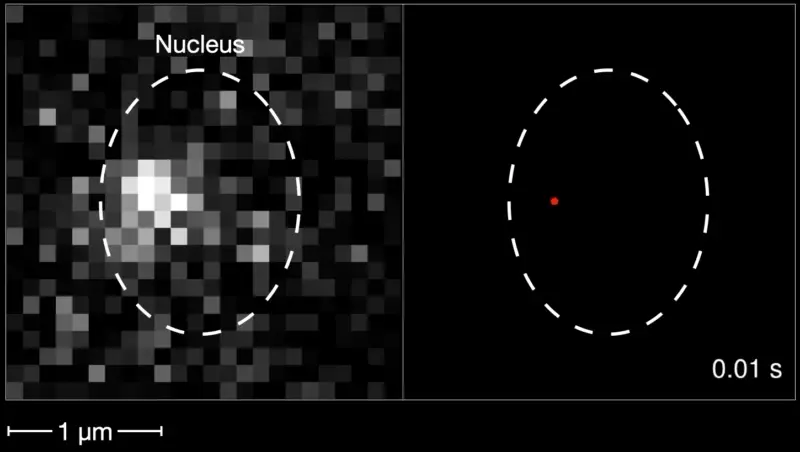
The general transcription factor TFIIH, required for transcription initiation, is present at ~2,000 molecules per yeast cell. A single molecule, imaged every 10 milliseconds, takes under 1 second to survey the entire nuclear space.
The Nguyen Lab uses superresolution microscopy to visualize and track genome regulators—chromatin-remodeling, epigenetic, and transcription machineries—in real time and at single-molecule resolution. Examining budding yeast and human cells in parallel, we aim to illuminate intranuclear traffic and establish fundamental principles underlying dynamic organization of eukaryotic transcription.
Select Publications
- Nguyen, V.Q., Ranjan, A., Liu, S., Tang, X., Ling, Y.H., Wisniewski, J., Mizuguchi, G., Li, K.Y., Jou, V., Zheng, Q., et al. (2021). Spatiotemporal coordination of transcription preinitiation complex assembly in live cells. Mol Cell 81, 3560-3575.e6
- Jeronimo, C., Angel, A., Nguyen, V.Q., Kim, J.M., Poitras, C., Lambert, E., Collin, P., Mellor, J., Wu, C., and Robert, F. (2021). FACT is recruited to the +1 nucleosome of transcribed genes and spreads in a Chd1-dependent manner. Mol Cell 81, 3542-3559.e11.
- Ranjan, A., Nguyen, V.Q., Liu, S., Wisniewski, J., Kim, J.M., Tang, X., Mizuguchi, G., Elalaoui, E., Nickels, T.J., Jou, V., et al. (2020). Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A.Z eviction. Elife 9, e55667.
- Nguyen, V.Q., Ranjan, A., Stengel, F., Wei, D., Aebersold, R., Wu, C., and Leschziner, A.E. (2013). Molecular Architecture of the ATP-Dependent Chromatin-Remodeling Complex SWR1. Cell 154, 1220–1231.
Biography
Vu Q. Nguyen immigrated from Vietnam in 1999 and attended SoCal's La Quinta High School, followed by three years at Orange Coast College. He then transferred to UC Berkeley and obtained his undergraduate degree in 2007. As an NSF predoctoral fellow, he studied structural biology using cryo-electron microscopy with Andres Leschziner at Harvard, receiving his Ph.D. in 2014. As a Damon Runyon postdoctoral fellow, he acquired expertise in live-cell, single-molecule microscopy with Carl Wu at HHMI Janelia and Johns Hopkins. In 2023, he returned to SoCal as Assistant Professor in Molecular Biology at UCSD.