New Insights on Cell Transport Motor Reveal a Surprise of Acceleration
Protein prompts speed as well as slowing for mechanism implicated in a range of diseases
September 7, 2017
By Mario Aguilera
Down at the level of individual molecules, cells are vast expanses of space. Traversing from one end of a cell to another can take an average protein several minutes. Other components clock in at hours… or even months in some cases.
In order for cells to develop and function normally, a system somewhat like railroad tracks and engines helps transport the cell’s cargo and get things where they need to be.
Groups led by Samara Reck-Peterson and Andres Leschziner at the University of California San Diego have made a surprising discovery about an essential cellular engine known as “dynein,” which transports cellular components on “microtubule” tracks to the right place at the right time and also plays a key role in cell division and neurodevelopment. Lis1, a protein that controls—or regulates—dynein motion was established to function as a dynein brake based on research the Reck-Peterson lab published in 2012. Research published September 7, 2017 in the journal Cell shows that was only half the story.
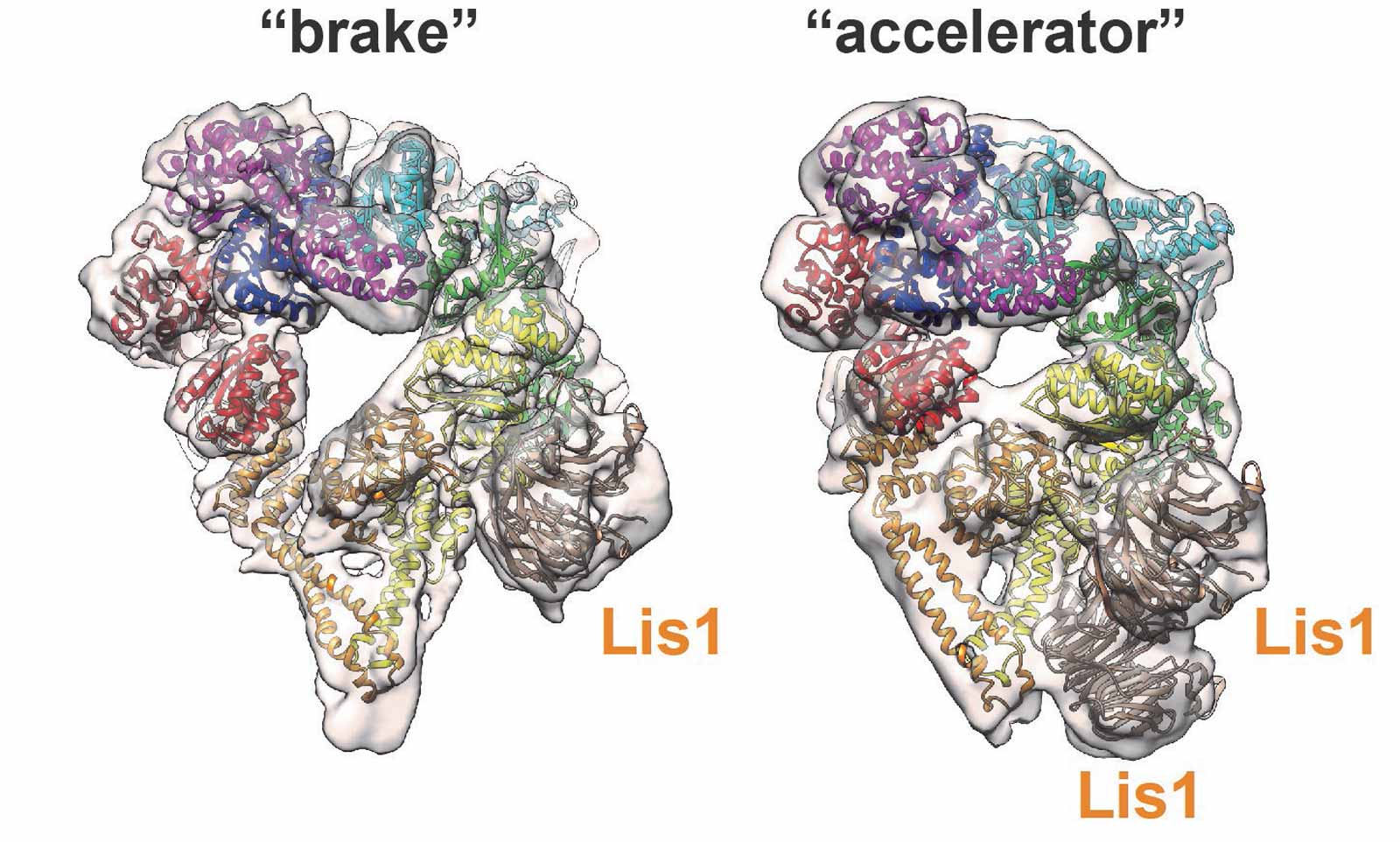
The 3D structures of dynein and Lis1, with Lis1 acting as a “brake” (left, one Lis1 attached to dynein) and “accelerator” (right, two Lis1s attached to dynein). The figure also illustrates how dynein changes depending on how many Lis1s are attached, from a more open appearance (left) to a more compact one (right).
The new study led by Morgan DeSantis, Michael Cianfrocco and Zaw Htet provides evidence that Lis1 can also help dynein step on the gas, in addition to pumping the brakes.
“Dynein is critical for normal development and mutations of proteins involved in dynein-mediated transport cause a range of neurodevelopment and neurodegenerative diseases,” said co-senior author Samara Reck-Peterson, a professor of Cell and Developmental Biology in UC San Diego’s Division of Biological Sciences and in the Department of Cellular and Molecular Medicine at UC San Diego School of Medicine. “The new study helps us understand how dynein is regulated in cells and why defects in this process could lead to disease.”
Mutations involving dynein dysfunction cause the brain developmental disease lissencephaly and the neurodegenerative diseases spinal muscle atrophy with lower extremity predominance and early onset Parkinson’s (Perry Syndrome) disease.
Using a combination of techniques, including 3-D structural analysis, to peer inside dynein’s engine, the researchers found that Lis1 can interact with dynein in two different ways. The new evidence showed that in one arrangement Lis1 weakens dynein’s drag along microtubule tracks, ultimately freeing it to move more quickly down the line.
“This is truly a case where seeing was believing” said co-senior author Andres Leschziner, a professor of molecular biology in UC San Diego’s Division of Biological Sciences and in the Department of Cellular and Molecular Medicine at UC San Diego School of Medicine. “It is very unusual for one protein to make another protein do opposite things, like braking or accelerating, so it was essential for us to do the structural analysis and directly see how Lis1 does this to dynein.”
Reck-Peterson and Leschziner say the researchers are now focusing on understanding how cells regulate Lis1 to cause it to act as either a brake or an accelerator at a given place and time.
Phuoc Tien Tran is also a coauthor of the paper.
The research included use of the Extreme Science and Engineering Discovery Environment for computing allocations (MCB160079 and MCB140257), supported by the National Science Foundation (NSF) (ACI-1548562). Support also included the Jane Coffin Childs Memorial Fund (61-1552-T), the Damon Runyon Cancer Research Foundation (2171-13), an NSF graduate research fellowship (DGE1144152), National Institutes of Health (R01GM107214), and the Howard Hughes Medical Institute and the Simons Foundation.
